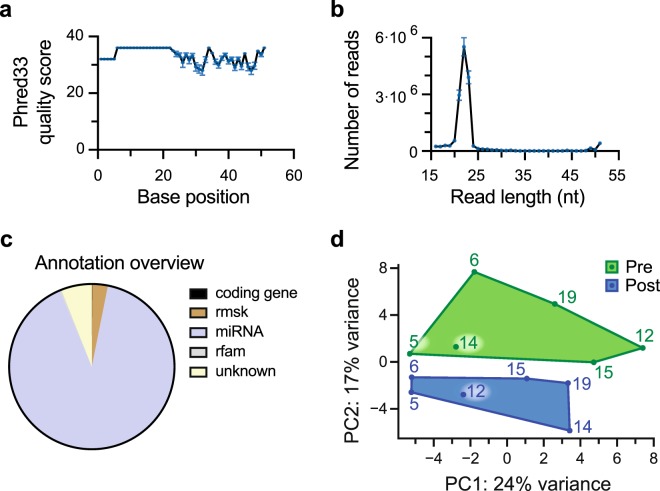
Figure 2.
Next generation sequencing of small non-coding RNAs purified from EVs. The quality score assigned to each base position after quality and adaptor trimming. (a) The read length distribution (b) shows a peak from 20–25 nucleotides. Annotation overview of reads (c), with approximately 80% of the reads mapping to miRNAs. PCA plot based on the counts of all detected miRNAs. (d) A green (pre-samples) and blue (post-samples) polygon denotes the smallest space to contain the pre and post samples, respectively. Rmsk: RepeatMasker, rfam: Database of functional non-coding RNA families.