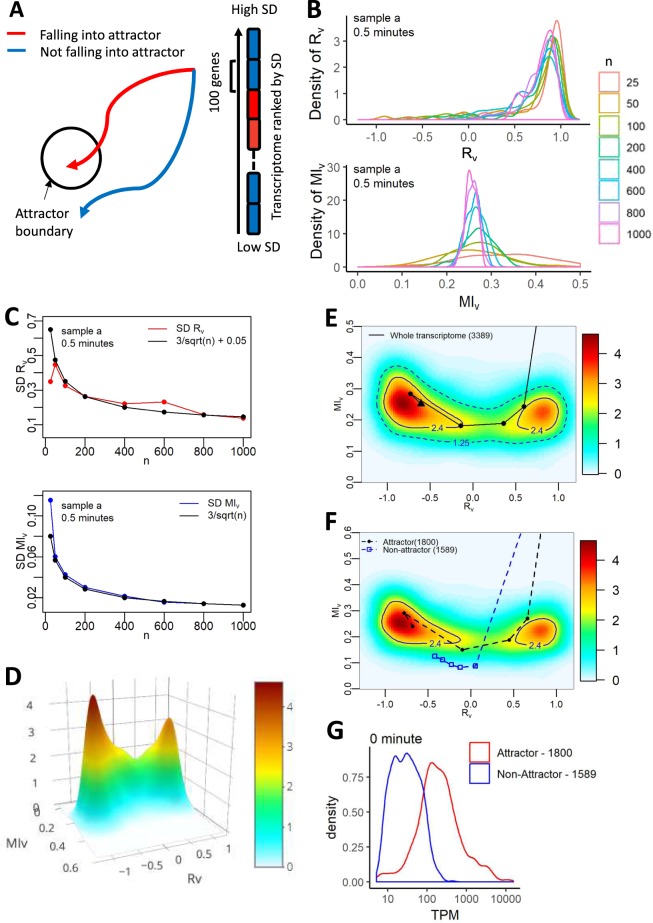
Figure 3.
Attractor landscape by probability density distributions of correlations, transcriptomic elements and attractor genes. (A) Schematic trajectories for transcriptomic elements falling into attractor (red) and not falling into attractor (blue). (B) Distribution and (C) Standard deviation of Rv (top panel) and MIv (bottom panel) with different transcriptomic element size (denoted as n) of replicate a at 0.5 minutes. Distribution of Rv and MIv for ensembles of n randomly chosen genes (n = 25, 50, 100, 200, 400, 600, 800, 1000) were generated with 100 repeats. Standard deviation of Rv and MIv decreases as n increases (except for when n = 25 for Rv), and follows + c law. See Fig. S3A,B for other time points. (D) 3D plot of the superimposition of the probability distribution (SPD) of Rv and MIv over all time points for the whole transcriptome. SPDs were estimated by getting Rv and MIv values of 100 randomly chosen genes for 100 times, using two-dimensional kernel density estimation. (E) Trajectory of whole genome (3389 genes) falling within attractor boundary (solid contour line) overlaid on SPD of whole transcriptome Rv and MIv. The trajectory was generated by averaging 100 trajectories of 100 randomly chosen genes from the pool of 3389 genes. (F) Trajectories of cumulative attractor (1800), and non-attractor (1589) genes overlaid on SPD of Rv and MIv for whole transcriptome. (G) Distribution of expression level for attractor (red), and non-attractor (blue) genes at representative t = 0. See Fig. S3D for other time points.