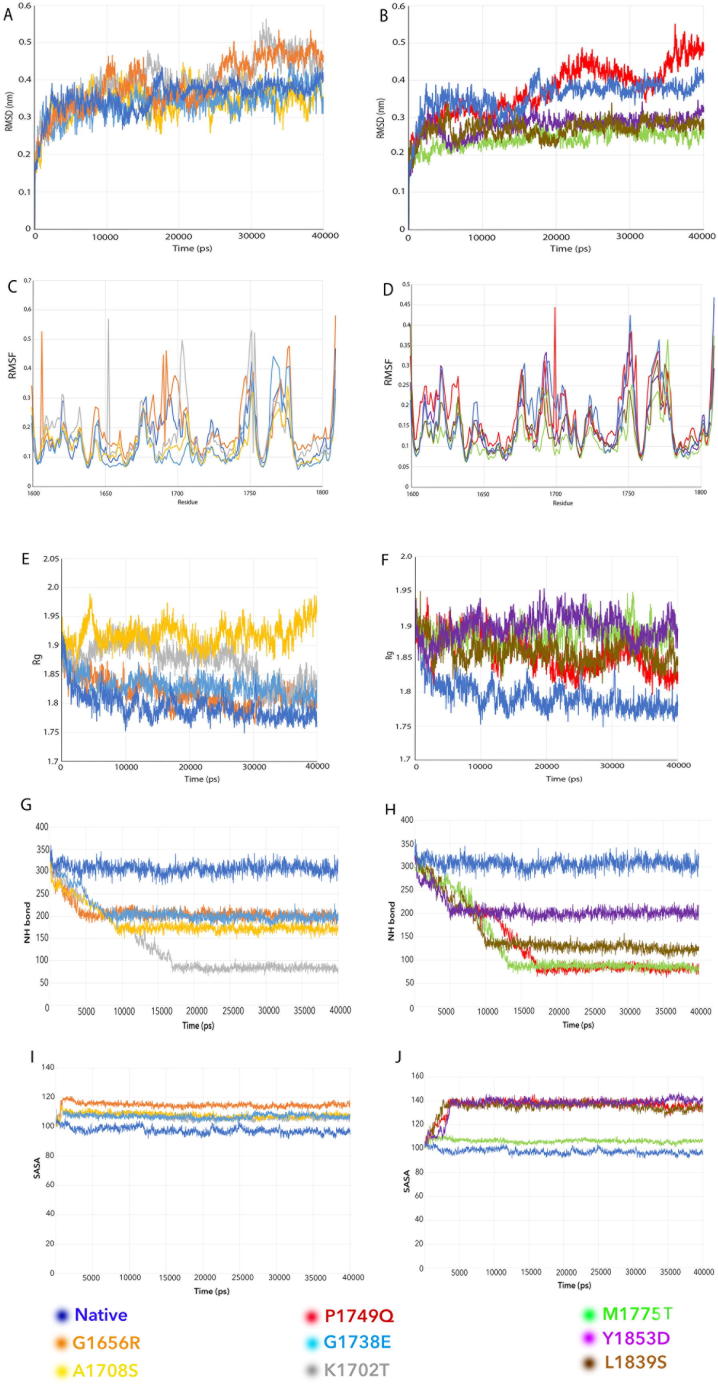
Fig. 2.
Examples of VUS classification by individual MDS programs. The x-axis represents the time period in each program. A, B. RMSD. The results show the RMSD of native and mutant protein complexes for a time period of 40 ns. The y-axis represents the RMSD for the native and variants structures. A. Native, G1738E, A1708S, G1656R, K1702T; B. Native, P1749Q, M1775T, L1839S, Y1853D. C, D. RMSF. The results show the RMSF profile of native and mutant protein complexes for a time period of 40 ns. The y-axis represents the RMSF score for native and variant structures. C. Native, G1738E, A1708S, G1656R, K1702T; D. Native, P1749Q, M1775T, L1839S, Y1853D. E, F. Rg. The results show the Rg profile of native and mutant protein complexes for a time period of 40 ns. The y-axis represents the Rg score for native and variant structures. E: Native, A1708S, G1738E, G1656R, K1702T; F: Native, P1749Q, M1775T, Y1853D, L1839S. G, H. NH-bond. The results show the number of intermolecular hydrogen bonds in native and mutant protein structures for a time period of 40 ns. The y-axis represents the no. of H bond for the native and variant structures. G: Native, A1708S, G1738E, G1656R, K1702T; H: Native, P1749Q, M1775T, Y1853D, L1839S. I, J. SASA. The results show the SASA profile of the native and mutant protein complexes for a time period of 40 ns. The y-axis represents the area in nm2. I. Native, G1738E, K1702T, G1656R, A1708S: J. Native, Y1853D, P1749Q, M1775T, L1839S.