Figure 1.
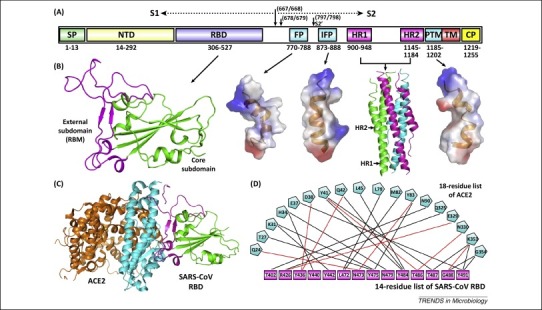
Severe acute respiratory syndrome coronavirus (SARS-CoV) spike features. (A) Schematic representation of the SARS-CoV spike protein (S). The individual components of S that were either experimentally characterized in previous studies – including receptor-binding domain (RBD), fusion peptide (FP), internal fusion peptide (IFP), heptad repeat 1/2 (HR1/2), and pretransmembrane domain (PTM) 13, 27, 35 – or are based on bioinformatics analyses, for example, N-terminal domain (NTD), are marked with the boundary-residue numbers listed below. The S1/S2 cleavage sites and the S2’-recognition site are highlighted. Other abbreviations: SP, signal peptide; TM, transmembrane domain; and CP, cytoplasmic domain. (B) Atomic structures of SARS-CoV spike RBD, FP, IFP, HR1/HR2 complex, and PTM (from left to right). The crystal structures of RBD (core subdomain in green and external subdomain in magenta) and the six-helix bundle fusion core (consisting of three HR1/HR2 helical hairpins in green, cyan, and magenta, respectively) are shown as ribbons, while the solution NMR structures of FP, IFP, and PTM are contoured using the electrostatic surface. (C) The complex structure between SARS-CoV RBD and its receptor ACE2. The core and external subdomains of RBD and the N- and C-terminal lobes of ACE2 are colored green, magenta, cyan, and orange, respectively. (D) The amino acid interactions at the RBD–ACE2 interface. According to a previous study [13], this binding network involves at least 18 residues in the receptor and 14 residues in SARS-CoV RBD, which are listed and connected with solid lines. Black lines indicate van der Waals contacts, and red lines represent H-bond or salt-bridge interactions.
