Abstract
A new insect nidovirus (named Yichang virus) from the family Mesoniviridae was isolated, identified, and characterized from Culex mosquitoes in Hubei, China. Results showed a high number of viral RNA copies (up to 1011 copies/ml) within 48 h in C6/36 cells. In addition, the titers of the Yichang virus reached maximal levels of 107 PFU/mL at 6 d post-infection (dpi). The virus produced moderate cytopathic effects when the multiplicity of infection ranged from 0.001–0.1 at 6 dpi, but did not replicate in mammalian cells. Under electron microscopy, the virion of the Yichang virus appeared as spherical particles with diameters of ∼80 nm and large club-shaped projections. Although subsequent genomic sequence analysis revealed that the Yichang virus had similar protein patterns as those of other mesoniviruses, the nucleotide acids shared less than 20% BLAST query coverage with known viruses in the family Mesoniviridae, and showed a maximum sequence identity of 67% for RNA-dependent RNA polymerase (RdRp). The putative protein sequences showed slightly higher identity (28%–68%), and the most conserved domain was RdRp. Based on the phylogenetic and pairwise evolutionary distance analyses, the Yichang virus should be considered a new species belonging to a currently unassigned genus within the family Mesoniviridae.
Keywords: Nidoviruses, Mesoniviridae, Yichang virus, Phylogeny, Taxonomic
1. Introduction
The RNA viruses within the order Nidovirales, which possess a linear single-stranded positive-sense RNA genome with a 5′ cap structure and 3′ poly(A) tail, are a genetically diverse group consisting of four families: Arteriviridae, Coronaviridae, Roniviridae, and the recently established Mesoniviridae (Lauber et al., 2012). Nidoviruses consist of approximately 80 virus species with a wide variety of hosts, ranging from crustaceans to mammals (Adams et al., 2016, Cowley and W.P., 2008, De Groot et al., 2012, Siddell et al., 2005). Members of Mesoniviridae have been isolated from various mosquito species in Africa (Côte d’Ivoire), Asia (Vietnam, Thailand, Korea, Indonesia, and China), Australia (Northern Territory), and North America (USA) (Hang et al., 2016, Kuwata et al., 2013, Liu et al., 2013, Nga et al., 2011, Thuy et al., 2013, Vasilakis et al., 2014, Warrilow et al., 2014, Zirkel et al., 2011, Zirkel et al., 2013), thus suggesting a wide geographic distribution. To date, however, only one mesonivirus, the Nam Dinh virus (NDiV), has been reported in China (Zhou et al., 2017). Based on phylogenetic and DEmARC (DivErsity pArtitioning by hieRarchical Clustering) analyses using either replicase non-structural proteins or large subsets of replicase non-structural polyproteins (3C-like proteinase (3CLpro) to O-methyltransferase (OMT) domains), Mesoniviridae has been divided into the genus Alphamesonivirus and one unassigned genus (Approved Proposal of the Mesoniviridae Study Group of the International Committee on Taxonomy of Viruses, ICTV, see Adams et al., 2016) [http://ictvonline.org/virusTaxonomy.asp].
All mesoniviruses have similar genome organization and replication strategies. Until now, all described viruses within the family Mesoniviridae possess structural and genetic similarities to other members in the order Nidovirales. Despite these similarities, however, the hosts of Nidovirales viruses can vary depending on the virus family; for example, mesoniviruses are hosted by insects, coronaviruses by insects and vertebrates, arterviruses by vertebrates, and roniviruses by crustaceans.
In our prior surveillance of the virome in mosquitoes collected from Hubei, China, we detected reads/sequences with similarities unique to Mesoniviridae viruses, indicating the possible existence of a mesonivirus in the collected samples (unpublished data). In the present study, this novel mesonivirus, which shares many conserved domains but is still only distantly related to other members of Mesoniviridae, was isolated from Culex mosquitoes. The virus morphology, growth properties, host range, genome organization, and putative proteins were determined. It is suggested that the virus is a new species (named Yichang virus) of Mesoniviridae. This study will help open new perspectives in and further our understanding of the family Mesoniviridae.
2. Materials and methods
2.1. Mosquito collection
From July to September 2014, a total of 10,101 adult female mosquitoes were collected and divided into 212 pools. The mosquitoes were classified into four species: Culex tritaeniorhynchus, Anopheles sinensis, Armigeres subalbatus, and Culex fatigans. They were captured in sites adjacent to human settlements in seven cities (Yichang, Wuhan, Qianjiang, Macheng, Huanggang, Xianning, and Ezhou) in Hubei, China. The 212 pools (each containing 50–100 mosquitoes) were created in accordance with the respective mosquito species collected and the sampling location. All collected mosquito samples were stored at −80 °C until analysis.
2.2. Sample preparation and nested-polymerase chain reaction (PCR) screening
Using sterile mortars and pestles, each mosquito pool was first treated with liquid nitrogen. The samples were then thoroughly ground, with 2 ml of RPMI medium then added for homogenization. A 0.22 μm membrane filter was used to remove debris and bacteria. RNA was extracted from the homogenized mosquito pool supernatants (n = 212) using the QIAmp Viral RNA Mini Kit (Qiagen, Germany). cDNA was synthesized using the GoScript™ Reverse Transcription system (Promega, USA) with random hexameric primers (R6) in accordance with the manufacturer’s instructions. Two pairs of nested primers were designed based on results obtained from previous Illumina sequencing (unpublished data), namely YichangV-F1 (5′-GCTAACCCACTTCGTCAA-3′) and YichangV-R1 (5′-AATACATCGGAGCCCAAC-3′) and YichangV-F2 (5′- TTTCATTTACCCTAACATCG-3′) and YichangV-R2 (5′- ATACATCGGAGCCCAACAAG-3′), respectively. Amplifications were carried out in a total volume of 25 μl consisting of 2 μl of template (cDNA), 0.25 μM of each primer, 12.5 μl of PrimerSTAR Max Premix 2 × (TaKaRa, Japan), and Milli-Q ddH2O to supplement the system. Amplification was performed using the following program: 98 °C/1 s, (98 °C/10 s, 52 °C/15 s, and 72 °C/5 s) × 30 cycles, after which 1.5 μl of the PCR product was transferred to a second tube with fresh reagents and inner primers YichangV-F2 and YichangV-R2.
Extensive nested-PCR was conducted to determine the prevalence rate of the Yichang virus in the 212 samples. Agarose gel electrophoresis was used to visualize the PCR products and DNA bands of interest were cut for sequencing.
2.3. Virus isolation, propagation, and purification
We infected C6/36 cells with the supernatant obtained from the positive samples determined by nested-PCR. Five serial passages were conducted to aid virus replication, with the viral RNA copies in each passage further determined by nested-PCR. To acquire pure virus stocks, we used the supernatant from the fifth passage to perform duplicate endpoint dilution titrations on the C6/36 cells.
To investigate viral replication in vertebrate cells, representative cells (African green monkey kidney [Vero], hamster kidney [BHK-21], and human embryonic kidney [293T]) were inoculated with the virus and incubated for 7 d at 37 °C. Five blind passages of cell culture supernatant were performed on fresh cells in 1:10 dilutions. Viral replication was then measured by real-time PCR and cytopathic effects (CPE) in each passage.
2.4. Genome sequencing
The viral RNA was extracted from the passaged virus, and then sent to The Beijing Genomics Institute (Illumina Miseq System) for sequencing. PCR was performed using specific primers (YichangV-gap-F1: TGTGCCGCCAGTTCTTAG/R1: TCAGGTCTGTGCGATGTA, YichangV-gap-F2: GCTAACCCACTTCGTCAA/R2: AATACATCGGAGCCCAAC), which were used to fill the gaps between the contigs. To acquire the terminal sequences, both 3′ and 5′ RACE were performed with high fidelity polymerase (Primer STAR Max Premix, TaKaRa, Japan). The 5′ RACE procedure was conducted using the 5′ SMARTer RACE Kit (TaKaRa, Japan), as per the manufacturer’s instructions. For the 3′ RACE procedure, reserve transcription using the 3′RACE reverse primer (3′RACE-Reverse: CGCACTCAGTCAGTTGCCGTTTTTTTTT) was performed to obtain an extended cDNA with a 20 nt barcode sequence, which was then used as the reverse primer (3′RACE-R: CGCACTCAGTCAGTTGCCG) for the second PCR assay, with the forward primer (3′RACE-F: ATAAGCATTGGGTCCAGAGTTGTCAAAC) designed based on the known sequence in the 3′-terminal.
The viral sequences were edited and assembled using SOAP denovo (http://soap.genomics.org.cn/soapdenovo.html) and verified by PCR using multiple primers. The full genome sequence was submitted to GenBank under accession number KY369959.
2.5. Virus growth kinetics
The C6/36 cells were infected with virus stocks purified through two endpoint dilutions, with MOIs of 0.1, 0.01, or 0.001, as described previously (Zirkel et al., 2013). Aliquots of the cell culture supernatant were removed at 12, 24, 36, 48, 60, and 72 h post-infection (hpi). RNA was then extracted using the Qiagen RNA Virus Mini Kit (Qiagen, Germany). Viral RNA copies of each sample collected at different time points were detected by real-time PCR. Amplifications were run on a MyiQ™2 Optics Module (Bio-Rad, USA) using a One Step SYBR PrimeScript™ PLUS RT-PCR Kit (TaKaRa, Japan). The standard reaction mixture contained 200 ng of sample RNA, 8 pmol of each primer, 10 μl of 2 × One Step SYBR RT-PCR buffer, 0.4 μl of PrimeScript plus RTase mix, 1.2 μl of TaKaRa Ex Tag HS mix, and 4.8 μl of RNase free dH2O to a total volume of 20 μl. All samples were analyzed with primers (RNA-dependent RNA polymerase (RdRp)-F: TTCGGTCGCATATACACAGC, RdRp-R: ACTTGTGGTTGGGATTGTCAG). Samples were run in triplicate to reduce the risk of false positive or negative results due to technical errors. Preincubation was performed for 5 min at 42 °C for reverse transcription, followed by 10 s at 95 °C then 40 cycles of 15 s at 95 °C for denaturation and 30 s at 60 °C for annealing and extension. Fluorescence was measured at the end of each cycle after completion of amplification (Langner et al., 2014).
2.6. Electron microscopy
Ultrastructural analysis of ultrathin sections of infected C6/36 cells followed that of previous research (Vasilakis et al., 2013) using a FEI Tecnai G20 transmission electron microscope (FEI Company, USA) at 200 kV.
The culture supernatants from infected cells showing CPE were centrifuged at 5000 × g for 10 min at 4 °C to remove cellular debris, followed by ultracentrifugation at 35,000 rpm (Type 70, Beckman) for 75 min at 4 °C to enrich the virus particles and second-round ultracentrifugation through a 20%–70% continuous sucrose gradient cushion at 39,000 rpm (SW40, Beckman) for 2 h at 4 °C. Further purification to collect the virus particles that formed a white band in the gradient was achieved via third-round ultracentrifugation at 40,000 rpm for 30 min at 4 °C to remove the sucrose. The virus pellets were re-suspended in 200 μl of PBS buffer overnight at 4 °C.
Purified virus particles were allowed to adhere to a Formvar carbon-coated copper grid for 10 min, and were then negatively stained with 2% phosphotungstic acid (PTA) with pH adjusted to 6.8 with 1 M KOH for 2–3 min. The particles were then examined using a Hitachi U8010 electron microscope (Japan).
2.7. Plaque assay
Aliquots of serial 10-fold dilutions of the virus in RPMI medium were inoculated with C6/36 cell monolayers in 6-well plates for 2 h. The cells were then covered with overlay medium, as performed in previous research (Vasilakis et al., 2013), and incubated at 28 °C for 6 d to allow for plaque development. Cells were subsequently stained with 2% crystal violet in 30% methanol for 5 min at room temperature. The plaques were manually counted and recorded as the number of plaques per ml of inoculum.
2.8. Protein analysis
Viral particles purified by ultracentrifugation in the sucrose gradient were lysed directly in a water-bath with 5 × SDS loading buffer [250 mM Tris-HCL, pH 6.8, 10% SDS (W/V), 0.5% bromophenol blue (W/V), 50% glycerol (W/V), 5% β-mercaptoethanol (W/V)] for 10 min at 100 °C. Subsequently, SDS-PAGE on a 12% polyacrylamide gel was employed to separate the viral protein bands, with the gel stained with Coomassie R250 for 3 h at room temperature. Multiple washing steps using washing buffer were performed until the redundant Coomassie dye was removed and distinct bands were visible.
Bioinformatics analysis of all mesoniviruses was conducted as described in reference (Zirkel et al., 2013) to characterize the putative proteins of the Yichang virus. Potential signal peptides, transmembrane domains (TMDs), and N-linked and O-linked glycosylation sites were identified using SignalP4.0 (Petersen et al., 2011) (http://www.cbs.dtu.dk/services/SignalP/), TMHMMv2.0 (Krogh et al., 2001) (http://www.cbs.dtu.dk/services/TMHMM/), and NetCGlyc (Julenius, 2007) (http://www.cbs.dtu.dk/services/NetCGlyc), respectively. Phosphorylation sites were identified using NetPhos 2.0 (Blom et al., 1999) (http://www.cbs.dtu.dk/services/NetPhos). The molecular weights and isoelectric points of the putative proteins were calculated with the pI/MW tool (Gasteiger et al., 2003) (http://web.expasy.org/compute_pi/).
2.9. Genome and phylogenetic analyses
Putative opening reading frame (ORF) organization was predicted by GeneMark (Besemer and Borodovsky, 2005) (http://exon.gatech.edu/GeneMark/heuristic_gmhmmp.cgi) and the Gene Finding in Viral Genomes function in the Softberry program (http://linux1.softberry.com/berry.phtml?topic=virus&group=programs&subgroup=gfindv). Conserved domains were predicted by HHpred (Soding et al., 2005) (https://toolkit.tuebingen.mpg.de/hhpred) and BLAST searching the National Center for Biotechnology Information (NCBI) database. Pseudoknot prediction was determined using the Ipknot server (Sato et al., 2011).
Amino acid sequences were aligned using CLUSTAL muscle (Edgar, 2004) modules with the UPGMB clustering method in the MEGA 6.06 software suite (Tamura et al., 2013). Alignment of the Yichang virus with representative members of the order Nidovirales focused on conserved regions, such as 3CLpro, helicase (Hel), RdRp (RNA dependent RNA polymerase), OMT, and N-methyltransferase (NMT). Phylogenetic analyses were conducted using MEGA 6.06 software with the Jones-Taylor-Thornton amino acid substitution model and uniform rates among sites with 1000 bootstrap replicates (Maximum Likelihood method).
3. Results
3.1. Prevalence and isolation
During sequencing analysis of the RNA extracted from the passaged virus obtained from mosquitoes collected in Hubei, China, three contigs were detected that matched most closely to the sequences of members of the Mesoniviridae family (specifically the Meno virus) (Zirkel et al., 2013).
After determining the whole genome here, the newly isolated virus was named “Yichang virus” based on the place of its isolation.
In the present study, the PCR results showed that the prevalence of the Yichang virus sequence in mosquito pools collected from Hubei was 16.5% (35 out of 212). The virus was most frequently detected in Culex mosquitoes, especially Culex quinquefasciatus (prevalence rate of 8.96%), and least frequently detected in Armigeres and Anopheles mosquitoes (1.4%) (Table S1). From the data available, this phenomenon might relate to the greater number of Culex mosquito samples in that cohort.
The supernatant from each PCR positive mosquito homogenate was inoculated on C6/36 monolayers for viral isolation. From the 35 positive samples obtained, only one virus strain was successfully isolated. This was likely due to competition with other viruses, insufficient quantity of the Yichang virus for infection, or destruction of the virus from multiple freezing and thawing cycles of the homogenates.
3.2. Genome organization
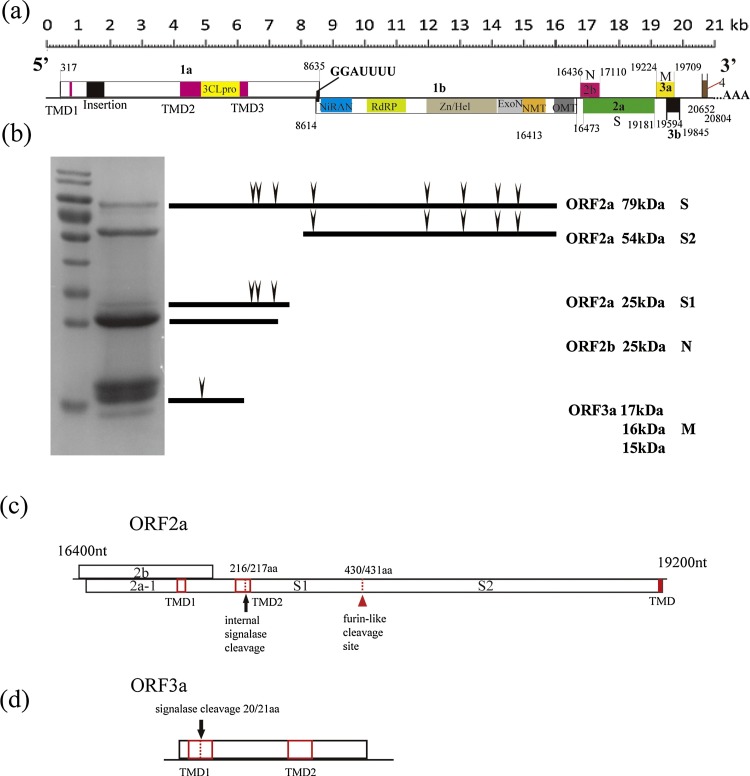
The entire genome of the Yichang virus was comprised of 20,893 nt [excluding the poly(A) tail]. This is the largest known genome in Mesoniviridae, with the second largest currently listed as the Karang Sari virus (20,777 nt, JKT-10701). Our results showed that the genomic organization was similar to that previously reported for viruses in this family (Vasilakis et al., 2014, Lehmann et al., 2015). The sizes of the 5′ and 3′ untranslated regions (UTRs) were 316 nt and 529 nt, respectively. We detected seven major ORFs and several small ORFs located behind ORF3, including ORF4, which are also conserved in all members of Mesoniviridae, except for the Meno virus (Fig. 1 a, Table 1 ) (Zirkel et al., 2013).
Fig. 1.
Yichang virus genome organization and structural proteins.
Schematic diagram of the organization of Yichang virus genome. Open reading frames (ORFs) are shown by boxes, with nucleotide positions indicated. Conserved domains are highlighted in colors. (b) SDS-PAGE analysis of the proteins for Yichang virus. Specific ORFs shown to encode the respective proteins are given on the right, together with the predicted molecular masses. Predicted N-linked glycosylation sites in the S and M proteins are indicated by triangles. (c) Transmembrane domains, signalase cleavage site and furin-like proteases cleavage site in the precursor ORF2a polypeptide. (d) Transmembrane domains and signalase cleavage site of precursor ORF3a polypeptide.
Table 1.
Six major open reading frames (ORFs) of the Yichang virus.
| ORF | Predicted protein | Genome position (nt) | Amino acid number (aa) | Molecular mass (kDa)a |
|---|---|---|---|---|
| 1a | Polyprotein 1a | 317–8635 | 2772 | 317.5 |
| 1b | Polyprotein 1b | 8614–16413 | 2599 | 301.4 |
| 2a | Spike protein | 16473–19181 | 902 | 103.5 |
| 2b | Nucleocapsid protein | 16436–17110 | 224 | 25.5 |
| 3a | Glycoprotein | 19224–19709 | 161 | 17.8 |
| 3b | Matrix protein | 19579–19908 | 109 | 12.7 |
Molecular mass value is predicted.
In addition, replicase domains identified in other viruses in the family (Nga et al., 2011, Vasilakis et al., 2013, Zirkel et al., 2013, Warrilow et al., 2014, Lehmann et al., 2015, Kuwata et al., 2013) were also predicted in the Yichang virus (Fig. 1a, Table 3 ). Amino acid similarities of the replicase domains and ORFs between Yichang and the other mesoniviruses ranged from 28% to 68%, exhibiting huge divergence (Table 2 ).
Table 3.
Mapping replicative protein domains on the Yichang virus genome.
| Range | Length | Name | Description | Target | Method | E-value |
|---|---|---|---|---|---|---|
| 21–43 aa (379–445 nt) | 23 aa | TMD1 | Transmembrane domain | ORF1a | TMHMM | |
| 1354–1510 aa (4376–4846 nt) | 157 aa | TMD2 | Transmembrane domain | ORF1a | TMHMM | |
| 1637–1788 aa (5225–5680 nt) | 151 aa | 3CLpro | Putative serine protease | ORF1a | Hhpred | 5.1E-07 |
| 1935–2022 aa (6122–6382 nt) | 87 aa | TMD3 | Transmembrane domain | ORF1a | TMHMM | |
| 8608–8614 nt | 7 nt | RFS | Ribosomal frame-shift site | ORF1a/ORF1b | ||
| 30-227aa (8701–9294 nt) | 198aa | NiRAN | Nidovirus RdRp-associated nucleotidyltransferase | ORF1b | Blastp | 6E-62 |
| 549–1017 aa (10258–11664 nt) | 469 aa | RdRp | RNA-dependent RNA polymerase | ORF1b | HHpred | 2.7E-38 |
| 1098–1808 aa (11905–14037 nt) | 711 aa | Zn/Hel | RNA ATP-dependent helicase NAM7 and zinc-binding domain | ORF1b | HHpred | 1.1E-38 |
| 1816–2116 aa (14059–14961 nt) | 301 aa | ExoN | 3′-5′exonuclease ERI1 | ORF1b | HHpred | 5.6E-27 |
| 2117–2354 aa (14962–15675 nt) | 238 aa | NMT | Guanine-N7 methyltransferase | ORF1b | HHpred | 2.3E-62 |
| 2406–2596 aa (15829–16401 nt) | 191 aa | OMT | 2′-O-methyltransferas-e SARS coronavirus | ORF1b | HHpred | 6.4E-13 |
Table 2.
Pairwise amino acid identities of replicase domains and complete open reading frames (ORFs) between the Yichang virus and other members of Mesoniviridae.
| Identity (aa) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus | Gene |
||||||||||||
| 3CLpro |
NiRAN |
RdRp |
ExoN |
Hel |
NMT |
OMT |
ORF1a |
ORF1ab |
ORF2a |
ORF2b |
ORF3a |
ORF3b |
|
| Yichang | |||||||||||||
| CavV | 48 | 48 | 66 | 42 | 51 | 39 | 43 | 32 | 43 | 39 | 48 | 47 | 30 |
| NDiV | 48 | 50 | 65 | 43 | 50 | 39 | 45 | 33 | 43 | 37 | 44 | 44 | 30 |
| KSaV | 48 | 51 | 66 | 44 | 49 | 49 | 51 | 32 | 50 | 37 | |||
| DKNV | 47 | 48 | 66 | 44 | 50 | 52 | 49 | 33 | 50 | 38 | 41 | 49 | 31 |
| CASV | 49 | 49 | 65 | 43 | 50 | 49 | 46 | 33 | 43 | 38 | 42 | 46 | 31 |
| HanaV | 41 | 49 | 65 | 42 | 51 | 42 | 45 | 33 | 43 | 38 | 42 | 47 | 30 |
| NseV | 33 | 51 | 65 | 44 | 51 | 41 | 43 | 32 | 43 | 36 | 42 | 48 | 28 |
| MenoV | 41 | 49 | 68 | 44 | 53 | 41 | 46 | 32 | 43 | 39 | 41 | 45 | 29 |
| MoumoV | 51 | 67 | 41 | 51 | 38 | 41 | 50 | ||||||
The putative TMDs in ORF2a-, 3a-, and 3b-encoded proteins were conserved among almost all mesoniviruses. Interestingly, the signalase cleavage site was wrapped inside the additional putative TMD of ORF2a and ORF3a (Fig. 1c, d); however, its exact impact on cleavage efficiency is not clear. Downstream of ORF3a/3b, several small ORFs (ranging in size from 25 to 50 aa) were predicted using the NCBI ORF Finder program, but no corresponding bands were discovered by SDS-PAGE analysis (Fig. 1b).
Detailed sequence analysis revealed that the insertion sequence of the Yichang virus included a large region of around 130 aa near the N-terminus of ORF1a (Fig. 1a), which is a highly variable region of different lengths unique to the mesoniviruses (Vasilakis et al., 2013). In addition, several short sequence stretches (ranging from 3 to 17 aa) were also present in ORF1a (data not shown), similar to those observed in the Bontag Baru, Karang Phet, and Karang Sari viruses (Vasilakis et al., 2014). Regarding the common sequence “SKRKGK” (Vasilakis et al., 2014), which was detected at the terminus of the largest insertion in the Yichang virus, we only found “KGK” conserved in the N-terminus, and no corresponding sequence at the C-terminus (Fig. S5). The larger number of short sequence stretches distributed in the ORF1a of the Yichang virus, in comparison with that of the other viruses in this family, might help explain its larger genome.
3.3. Phylogenetic analysis and species determination
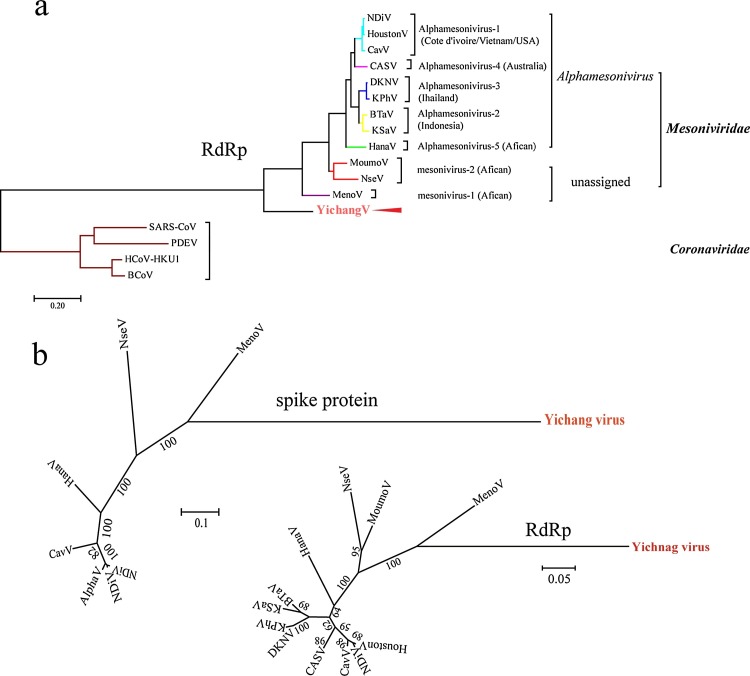
Phylogenetic relationships were constructed based on the amino acid alignments of ORF2a (whole S protein) and the highly-conserved domains within ORF1ab (3CLpro, RdRp, and Hel) (Fig. 2 ).
Fig. 2.
Phylogenetic relationships of mesoniviruses.
Phylogenetic trees of mesoniviruses and representative nidoviruses based on amino acid sequences of RdRp and spike proteins were conducted by the Maximum-likelihood (ML) method, a NJJ substitution matrix, and no distance correction in MEGA 6.06. Significance was tested by bootstrap analysis using 1000 resampling steps. The scale bars represent number of substitutions per amino acid position on average.
(a) Rooted based on RdRp protein and including roniviruses as outgroup.
(b) two unrooted of mesoniviruses and based on spike and RdRp proteins
The 3CLpro and Hel phylogenetic trees, which included many representative nidoviruses, consistently showed the Yichang virus to be clustered with the Mesoniviridae family. Further to this, the 3CLpro phylogenetic tree, which consisted of only mesoniviruses, indicated that the new virus should be classified into an unassigned group along with mesonivirus 1 and mesonivirus 2.
To clarify the phylogenetic relationship between the Yichang virus and other mesoniviruses, three radiation trees were determined by alignment of the 3CLpro, spike protein, and RdRp domains, which are conserved in this family. Our results illustrated that the Yichang virus was divergent from the other viruses. The Yichang virus was also clearly basal to the other viruses in the 3CLpro and Hel phylogenetic trees, suggesting that the Yichang virus is likely more ancient from an evolutionary perspective.
Although the phylogenetic trees exhibited highly similar topologies and strongly indicated that the Yichang virus should be clustered within the family Mesoniviridae, results also showed that the virus was still distinct from all other mesoniviruses (Fig. 2).
To further confirm the relationship between the Yichang virus and other mesoniviruses, pairwise evolutionary distances (PED) of the conserved protein domains within ORF1ab were calculated. Results indicated that the PED values between the Yichang virus and each previously described strain were all substantially greater (>0.7) than the suggested cutoff of 0.092 established for species demarcation (ICTV) and the 0.32 threshold recognized for genus separation. Thus, the Yichang virus should be considered as a unique species and classified to an unassigned group (Table 4 ).
Table 4.
Pairwise evolutionary distances (PED) using MEGA 6.06 software based on conserved domains in ORF1b.
| NseV | MenoV | HanaV | AlphaV | CASV | HouV | KPV | KSV | BTV | NdiV | CavV | Yichang | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NseV | ||||||||||||
| MenoV | 0.405 | |||||||||||
| HanaV | 0.351 | 0.399 | ||||||||||
| AlphaV | 0.337 | 0.387 | 0.168 | |||||||||
| CASV | 0.349 | 0.398 | 0.244 | 0.229 | ||||||||
| HouV | 0.335 | 0.384 | 0.169 | 0.009 | 0.229 | |||||||
| KPV | 0.349 | 0.398 | 0.198 | 0.157 | 0.237 | 0.156 | ||||||
| KSV | 0.361 | 0.411 | 0.223 | 0.180 | 0.266 | 0.180 | 0.154 | |||||
| BTV | 0.370 | 0.412 | 0.226 | 0.181 | 0.268 | 0.180 | 0.158 | 0.077 | ||||
| NdiV | 0.334 | 0.384 | 0.169 | 0.016 | 0.228 | 0.019 | 0.156 | 0.180 | 0.182 | |||
| CavV | 0.341 | 0.390 | 0.177 | 0.079 | 0.228 | 0.078 | 0.163 | 0.189 | 0.192 | 0.079 | ||
| Yichang | 0.733 | 0.701 | 0.710 | 0.711 | 0.719 | 0.709 | 0.704 | 0.714 | 0.709 | 0.709 | 0.707 |
NseV: Nse virus, MenoV: Meno virus, HanaV: Hana virus, AlphaV: Alphamesonivirus 1, CASV: Casuarina virus, NgeV: Ngewoton virus, HouV: Houston virus.
KPV: Kamphang Phet virus, KSV: Karang Sari virus, BTV: Bontang virus, NdiV: Nam Dinh virus, CavV: Cavally virus, Yichang: Yichang virus.
Alphamesonivirus 1 strain A12.2520/ROK/2012KU095838.1.
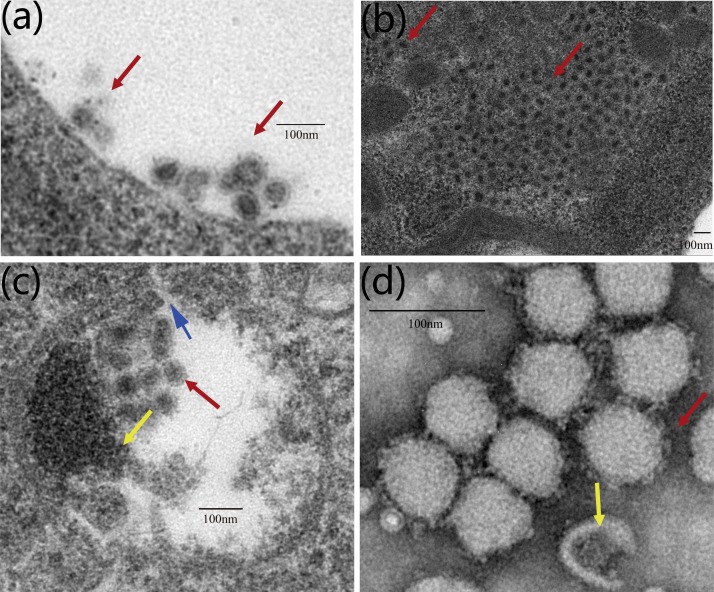
3.4. Virus morphology and growth
In the ultrathin sections of fixed infected C6/36 cells analyzed by transmission electron microscopy at 48, 72, and 96 hpi, mature virions with diameters of ∼80 nm were detected at the surface of the cell membrane or around the vesicles as either individual particles or groups (Fig. 3 a, b). This appearance was the same as that observed for other members in this family (Vasilakis et al., 2014). Vesicles were observed in the cytoplasm of infected cells, containing premature viral particles with diameters of ∼60 nm (Fig. 3c). Spike proteins protruded from the envelope by ∼10 nm (Fig. 3d). In comparison, the lengths of spike proteins in other members of Mesoniviridae range from 12 nm in the CavV (Zirkel et al., 2013), Hana, Nse, and Meno viruses, to 3–4 nm in the NDiV virus (Thuy et al., 2013) and 15 nm in the Casuarina virus (Warrilow et al., 2014) (Table S2).
Fig. 3.
Yichang virus replication and morphology as observed by transmission electron microscopy.
(a) and (b) Mature virions (red arrow) with ∼80 nm in diameter were situated at the surface of the cell membrane, or around the vesicles, either as individual particles or groups. (c) Ultrathin sections of C6/36 cells infected with Yichang virus, vesicles containing spherical, enveloped particles with diameter 70–80 nm (red arrow), particles lacking the envelope proteins with diameter 50–60 nm (yellow arrow),an opening or a pore in the vesicles which allow movement of the viral RNA from the vesicles to the cytosol (blue arrow). (d) Negative stained preparation of Yichang virus particles sedimented by ultracentrifugation, showing complete (red arrow) and incomplete (yellow arrow) virions. The length of the putative spikes on the virion is approximately 10 nm.
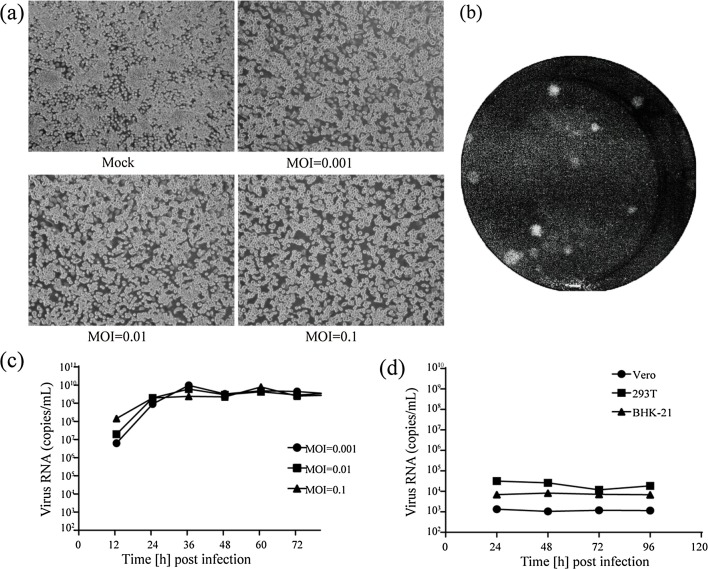
The purified Yichang virus induced mild CPE, manifesting in aggregation and detachment in the C6/36 cells at 6 d post-infection (dpi) (Fig. 4 a), but not in the cultured vertebrate cell lines (Vero, BHK-21, 293T) (Fig. 4d). The titers grown in the C6/36 cells reached maximal levels of 107 PFU/mL at 6 dpi, as determined by plaque assay (Fig. 4b). As shown in Fig. 4, the titers of the Yichang virus were slightly lower than those of the CavV, Hana, Nse, and Meno viruses calculated by TCID50 (Zirkel et al., 2013), and considerably lower than that of the Dak Nong virus (up to 1011 PFU/mL) measured by plaque assay (Kuwata et, al., 2013) (Table S2).
Fig. 4.
Yichang virus growth on insect and vertebrate cell lines.
(a) Cytopathic effect (CPE) was observed at 6 d postinfection in the infected C6/36 cells (right)(MOI = 0.1-0.001)compared to the mock (left). (b) Representative plaques of Yichang virus −infected C6/36 cells at 6 d postinfection. (c) The growth curve (MOI = 0.1–0.001) was measured by RT-PCR at 12–72 h postinfection.
(b) The growth curve measured by RT-PCR on three different vertebrate cell lines.
The viral RNA copies in the infected cells were detected by real-time PCR. An aliquot of cell culture supernatant was removed every 12 h for 3 d following infection. The amount of viral RNA increased to 109 RNA copies/mL within 24 h, and peaked at 1010 RNA copies/mL at 36 h with an MOI of 0.1–0.001, indicating a fast replication cycle (Fig. 4c). However, moderate CPE was observed at 6 dpi, which was much later than the observed peak in RNA copies (Fig. 4a).
3.5. Structural proteins
The SDS-PAGE analysis revealed that the Yichang virus was composed of at least seven major structural polypeptides: S (79 kDa), S2 (54 kDa), S1 (25 kDa), N (25 kDa), and M proteins (17, 16, and 15 kDa) (Fig. 1b).
Three of the polypeptides (S, S2, and S1), known as spike proteins, were deduced from ORF2a. The amino acids of these spike proteins were quite distinct from those of the other mesoniviruses, based on the low identity ranges of 36%–39% (Table 2). Bioinformatics analysis revealed that there were two post-translational cleavage sites, that is internal signalase and furin-type cleavage sites, in the spike glycoprotein precursor (Fig. 1c), which have also been observed in other mesoniviruses along with many conserved features. The internal signalase cleavage site in the Yichang virus was predicted by SignalP4.1 and was located at the site 213KAHA216/217YTRIN219, which is conserved at the “TRI” residue (Zirkel et al., 2013). The furin-type cleavage site was predicted by ProP1.0, and showed that the multibasic cleavage site 426SRSKR/WDSS434 (Yichang virus) was highly conserved among mesoniviruses (Zirkel et al., 2013) (Fig. S1).
Alignment of ORF2a showed that the Yichang virus had 12 conserved cysteine residues (Fig. S1) and four conserved potential N-glycosylation sites across all mesoniviruses (Vasilakis et al., 2014) (Fig. 1c, Table 5 ).
Table 5.
Putative proteins encoded by ORF2a.
| Proteins | Amino acid number (aa) | Predicted molecular mass (kDa) | Glycosylation site number |
|---|---|---|---|
| 2a-1 | 216 | 24.4 | 3 |
| S | 686 | 79 | 8 |
| S1 | 212 | 25.04 | 3 |
| S2 | 472 | 54 | 5 |
The nucleotide capsid protein (abbreviation N) was encoded by ORF2b without a TMD or N-glycosylation site (Fig. 1b).
The three remaining 17, 16, and 15 kDa proteins were matched on the putative M protein (membrane protein) encoded by ORF3a, which was predicted to contain a signal peptidase cleavage site 17YIYP/TDAT24, representing high amino acid diversity in this family (Fig. 1d, Fig. S3). Predictions of N-glycosylation sites for putative M using NetNGlyc exhibited only one site, which could not explain the three different sizes of membrane proteins observed by SDS-PAGE.
MUSCLE alignment of all available mesonivirus ORF3a protein sequences (Fig. S4) indicated that the signalase cleavage site was relatively variable.
3.6. Putative replicase polyprotein
The replicase domains in Mesoniviridae were also conserved in the Yichang virus (Fig. 1a, Table 3), and included three TMDs (TMD1, TMD2, and TMD3), a 3CLpro, a ribosomal frameshift site (RFS), a nidovirus RdRp-associated nucleotidyltransferase (NiRAN) (Lehmann et al., 2015), an RdRp, an RNA Hel, a 3′-5’exonuclease (ExoN), a zinc-binding domain (ZBD), a guanine-N7 methyltransferase, and a ribose-2′-OMT (Gosert et al., 2002, Nga et al., 2011, Snijder et al., 2003, Zirkel et al., 2013). Among these enzymes, the ExoN domain exhibits properties that are relevant for understanding the relationship between genome size and mutation rate in RNA viruses (Chen et al., 2007, Minskaia et al., 2006).
Comparative sequence analysis showed that the RdRp domain had the highest amino acid identity to homologous regions of the mesoniviruses (ranging from 65%–68%) (Table 2). In addition, the 3CLpro conserved domain of the Yichang virus was a cysteine protease with a chymotrypsin-like fold with a Cys (2968)-His (2879) catalytic dyad conserved in this family (Blanck et al., 2014). Research on the cleavage sites of 3CLpro has already shown 12 conserved sites in the ORF1ab, and substrate specificity of 3CLpro has shown that the Aln (N) residue is conserved in these cleavage sites (Blanck and Ziebuhr, 2016). These features were also predicted by multiple alignment, with seven of these sites found to be conserved in the Yichang virus (Fig. S2, S4.)
3.7. Ribosomal frame-shift site
Mesoniviruses usually have two RFSs located at the ORF1a/ORF1b and ORF3a/ORF3b overlaps, and the translation of ORF1b and ORF3b characteristically occurs through the RFS at a ‘slippery’ sequence to allow read-through synthesis of a polyprotein (pp1ab or pp3ab) (Brierley, 1995, Ziebuhr et al., 2000).
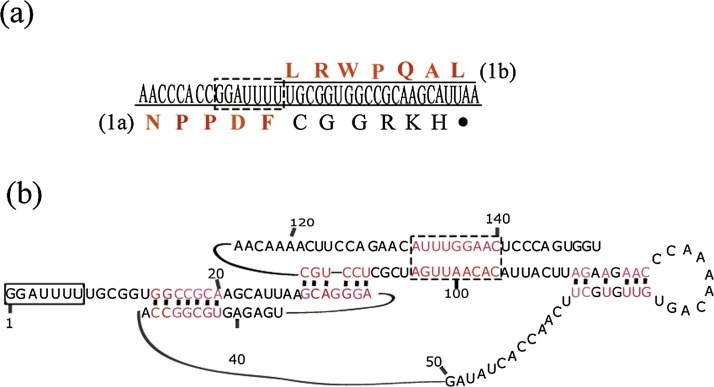
The translation of ORF1b is usually facilitated by an RNA pseudoknot in the sequence immediately downstream of the RFS. This functional pseudoknot structure is conserved in nidoviruses, which usually have four major conserved domains (Vasilakis et al., 2014). Therefore, alignment of all mesonivirus sequences extending 150 nt from the start of the ‘slippery sequence’ was conducted using Ipknot (Sato et al., 2011), which predicts the consensus secondary structure of the aligned sequences (Fig. 5 b). As shown in Fig. 5b, the Yichang virus only had three conserved domains in the pseudoknot region, and the complete RFS or RFS core sequences (CACUUUU) were not found in the ORF3a/ORF3b overlap region.
Fig. 5.
Predicted structures of ORF1a/ORF1b ribosomal frame-shift element (RFS) and RNA prseudoknot structure in the Yichang virus genome.
(a) The genomic nucleotide (middle) and amino acid sequences (lower) of ORF1a and ORF1b around their overlapping sites. (b) A conserved pseudoknot structure of RNA immediately downstream of slippery sequence (GGAUUUU) about 150nt which reduce ribosomal −1 frameshift (RFS) in 12 typical mesoniviruses.
4. Discussion
In this study, we isolated and identified a novel mosquito virus from Hubei province in China. This virus shared many common characteristics with other mesoniviruses. Based on the phylogenetic and pairwise evolutionary distance analyses, we propose that the Yichang virus is a new species that likely belongs to an unassigned genus within the family Mesoniviridae. This is the first time a new species within Mesoniviridae has been reported in China.
Significant divergence was expressed in the nucleotide and amino acid similarities. To date, the genome sequence of the Yichang virus reported here is unique among other mesoniviruses, although it still possesses many of the conserved features of this family. This novel virus could provide new perspectives for research on the evolutionary relationships of mesoniviruses, including the nidoviruses.
Our study showed that the Yichang virus has the largest genome size in the family Mesoniviridae. Detailed analysis and comparisons of genome organization revealed some notable differences, such as the presence of block insertions of up to 390 nt (130 aa) in the 5′ terminal quadrant of ORF1a compared with the CavV, Hana, Houston, Meno, and Casuarina viruses. The block insertion region was flanked by TMD1 and TMD2 and no conserved domains were found. In the same area, the Dak Nong (DKNV, 61 aa), Meno (13 aa), Kamgphang Phet (66 aa), Karang Sari (196 aa), and Bontag Baru viruses (193 aa) exhibit similar features (Vasilakis et al., 2014, Kuwata et al., 2013), but share low amino acid similarities. For example, the common sequence “SKRKGK” has been detected in BTaV, KsaV, and KPhV at both the N- and C-terminuses and in DKNV on the N-terminus, but only partially (“KGK”) found at the N-terminus in the Yichang virus, and not located in the Meno virus at all. The imperfectly repeated sequences (KAEE and RKGN) within each insertion region, which suggest possible origin through recombination or sequence duplication events, were not found in the Yichang virus (Vasilakis et al., 2014). Moreover, this insertion feature is only found in the Asian strains, although its function remains unknown. Previous analysis of the evolution of nidovirus genomes concluded that genome expansion occurred in a wave-like fashion, in which the three major coding regions (OFR1b, ORF1a, and 3′ORFs) expanded consecutively in a hierarchy that reflects the roles of their encoded proteins in the virus replication cycle (Lauber et al., 2013). The newly isolated Yichang virus provides evidence that nidoviruses have an inherent capacity for genome expansion, most likely associated with the transitional retention of sequences that serve as a resource for the evolution of new functions.
As shown in earlier research (Vasilakis et al., 2014), the conserved pseudoknot structure is responsible for the activation of the −1 ribosomal frame-shift (as observed in our study, Fig. 5a), rather than the stem-loop of the immediate downstream sequence of the RFS. The predicted structure of the Yichang virus featured only three relatively short regions of complementary pseudoknot domains instead of four, as previously reported (Vasilakis et al., 2014); therefore, we speculated that these three regions are important due to their strong conservation. It should be noted that a common ‘slippery’ sequence (CACUUUU) found in other members of Mesoniviridae was not detected in the Yichang virus, and the Yichang virus sequence could not be read through in this region. Thus, the mechanism of mesonivirus gene expression is still unclear.
Based on the ICTV demarcation criteria, if a virus has a PED value of more than 0.092 but less than 0.311 against other viruses within a genus, it should be classified as a new species. As the Yichang virus had PED values of more than 0.311 among the viruses within the genus Alphamesonivirus 1 and the other two unassigned genera it should be defined as a new species. In the 3CLpro and Hel phylogenetic trees, the Yichang virus displayed an obviously basal position compared with that of the other viruses, suggesting a more ancient origin from an evolutionary perspective.
Extensive phylogenetic analysis of mesonivirus relationships suggests that the Meno virus might represent a sister taxon to all other mesoniviruses (Zirkel et al., 2013). Here, the 3CLpro phylogenetic tree indicated that the Yichang virus had a close relationship with the Meno virus.
Our phylogenetic analyses clearly showed that the Yichang virus formed a new branch in this family that was distant from known mesoniviruses. Thus, the Yichang virus is proposed as a new species not belonging to the Alphamesonivirus genus, which could help in future analysis of the evolutionary patterns of Mesoniviridae.
Acknowledgements
We thank the Hubei Provincial Center for Disease Control and Prevention for help with sample collection. This work was supported by the Ministry of Science and Technology of China (Science and Technology Basic Work Program 2013FY113500).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2017.08.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Adams M.J., Lefkowitz E.J., King A.M., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch. Virol. 2016;161(10):2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33(Web Server issue):W451–454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck S., Ziebuhr J. Proteolytic processing of mesonivirus replicase polyproteins by the viral 3C-like protease. J. Gen. Virol. 2016;97(6):1439–1445. doi: 10.1099/jgv.0.000458. [DOI] [PubMed] [Google Scholar]
- Blanck S., Stinn A., Tsiklauri L., Zirkel F., Junglen S., Ziebuhr J. Characterization of an alphamesonivirus 3C-like protease defines a special group of nidovirus main proteases. J. Virol. 2014;88(23):13747–13758. doi: 10.1128/JVI.02040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76(Pt 8):1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Chen P., Jiang M., Hu T., Liu Q., Chen X.S., Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 2007;40(5):649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., W.P . In: Nidoviruses. Perlman S.G.T., Snijder E.J., editors. ASM Press; Washington DC: 2008. Molecular biology and pathogenesis of roniviruses; pp. 361–377. [Google Scholar]
- De Groot, R.J., C.J., Enjuanes, L., Faaberg, K.S., Perlman, S., Rottier, P.J.M., Snijder, E.J., Ziebuhr, J., Gorbalenya, A.E., 2012. Order Nidovirales. In: King, A.M.Q., A.M., Carstens, E.B., Lefkowitz, E.J., (Ed.), Virus taxonomy; Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier London, pp. 785–795.
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J., Klein T.A., Kim H.C., Yang Y., Jima D.D., Richardson J.H., Jarman R.G. Genome sequences of five arboviruses in field-Captured mosquitoes in a unique rural environment of South Korea. Genome Announc. 2016;4(1) doi: 10.1128/genomeA.01644-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17(8):868–876. doi: 10.1093/glycob/cwm050. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuwata R., Satho T., Isawa H., Yen N.T., Phong T.V., Nga P.T., Kurashige T., Hiramatsu Y., Fukumitsu Y., Hoshino K., Sasaki T., Kobayashi M., Mizutani T., Sawabe K. Characterization of Dak Nong virus, an insect nidovirus isolated from Culex mosquitoes in Vietnam. Arch. Virol. 2013;158(11):2273–2284. doi: 10.1007/s00705-013-1741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P.T., Morita K., Snijder E.J., Gorbalenya A.E. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157(8):1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9(7):e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Lin L., Zhou J.M., Chen Y.J., Zhang Q.W., Wang D.Q., Li J.M., Jin Y.J. [Identification of nam dinh virus in China] Bing Du Xue Bao. 2013;29(1):1–6. [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3’-5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Sato K., Kato Y., Hamada M., Akutsu T., Asai K. Ipknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27(13):i85–93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell, S., Ziebuhr, J., Snijder, EJ., 2005. Coronaviruses, Toroviruses and Arteriviruses, edited by (ed.), i.F.C. Topley and Wilson’s Microbiology and Microbial Infections, Edward Arnold London.
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy N.T., Huy T.Q., Nga P.T., Morita K., Dunia I., Benedetti L. A new nidovirus (NamDinh virus NdiV): Its ultrastructural characterization in the C6/36 mosquito cell line. Virology. 2013;444(1–2):337–342. doi: 10.1016/j.virol.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Forrester N.L., Palacios G., Nasar F., Savji N., Rossi S.L., Guzman H., Wood T.G., Popov V., Gorchakov R., Gonzalez A.V., Haddow A.D., Watts D.M., da Rosa A.P., Weaver S.C., Lipkin W.I., Tesh R.B. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013;87(5):2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Guzman H., Firth C., Forrester N.L., Widen S.G., Wood T.G., Rossi S.L., Ghedin E., Popov V., Blasdell K.R., Walker P.J., Tesh R.B. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 2014;11:97. doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D., Watterson D., Hall R.A., Davis S.S., Weir R., Kurucz N., Whelan P., Allcock R., Hall-Mendelin S., O’Brien C.A., Hobson-Peters J. A new species of mesonivirus from the northern territory, Australia. PLoS One. 2014;9(3):e91103. doi: 10.1371/journal.pone.0091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Jin Y., Chen Y., Li J., Zhang Q., Xie X., Gan L., Liu Q. Complete genomic and ultrastructural analysis of a nam dinh virus isolated from culex pipiens quinquefasciatus in China. Sci. Rep. 2017;7(1):271. doi: 10.1038/s41598-017-00340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Zirkel F., Kurth A., Quan P.L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C., Junglen S. An insect nidovirus emerging from a primary tropical rainforest. mBio. 2011;2(3):e00077–00011. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Roth H., Kurth A., Drosten C., Ziebuhr J., Junglen S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013;87(11):6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.