Highlights
-
•
An improved molecular diagnostics tool that utilizes CRISPR/dCas9-mediated biosensor.
-
•
Couples a nuclease inactivated Cas9 (dCas9) and single microring resonator biosensor.
-
•
Achieved single molecule sensitivity for the detection of ST (0.54 aM) and SFTS (0.63 aM).
-
•
CRISPR/dCas9-mediated biosensor can detect pathogens in 20 min.
Keywords: Clinical diagnosis, CRISPR/Cas system, Nuclease inactivated Cas9, SMR biosensor, In vitro, Diagnostics
Abstract
Rapid and highly sensitive detection of biomolecules is greatly needed for pathogen diagnosis in clinical samples, but the method needs to be significantly improved in terms of sensitivity and specificity for actual use in clinical settings. Here, we report the development of an improved molecular diagnostics tool that utilizes CRISPR/dCas9-mediated biosensor that couples a nuclease inactivated Cas9 (dCas9) and single microring resonator biosensor, enables label-free and real-time detection of pathogenic DNA and RNA. We addressed the clinical utility of this CRISPR/dCas9-mediated biosensor in tick-borne illnesses including scrub typhus (ST) and severe fever with thrombocytopenia syndrome (SFTS), whose clinical presentations are too similar to be easily differentiated. By using CRISPR/dCas9-mediated biosensor, we achieved single molecule sensitivity for the detection of ST (0.54 aM) and SFTS (0.63 aM); this detection sensitivity is 100 times more sensitive than that of RT-PCR assay. Finally, CRISPR/dCas9-mediated biosensor was able to clearly distinguish between ST and SFTS in serum samples within 20 min. We believe that CRISPR/dCas9-mediated biosensor will be useful for rapid and accurate molecular diagnostic tool that is suitable for immediate clinical applications.
1. Introduction
Due to the recent increase in the occurrence of emerging infectious diseases such as Zika, Ebola, and MERS-CoV, advances have been made for fast detection of target pathogens [[1], [2], [3]]. To control the spreading of pathogens from one community to another and to provide accurate treatment to patients, correct diagnosis of pathogen in first-line diagnostic places including clinics is crucial. Although many different diagnostic approaches have been introduced, development of a rapid and high sensitive approach for clinical diagnosis still remains a challenge.
Rapid diagnostic testing (RDT) based on antibody detection for infectious diseases produces results within an hour [4,5]. However, the detection sensitivity of RDT is lower than that of sequence-based nucleic acid amplification methods such as PCR [[6], [7], [8]]. Hence, sequence-based nucleic acid testing has many applications in clinical fields such as diagnosis and epidemiology. However, those approaches are limited in that they are long processing time, high-cost, and relatively less sensitive for clinical use [9]. Over the past decade, biosensors combined with electrical, electrochemical, and optical techniques have been developed to improve detection sensitivity [10].
Recently the nucleic acid amplification and detection technologies that enable rapid, simple, and sensitive detection of biomolecules have been developed. The isothermal solid-phase DNA amplification and detection (iSAD) to simultaneous amplify and detect DNA molecules on silicon microring resonator (SMR) biosensor, and the isothermal one-step RNA amplification and detection (iROAD) allows for amplification and detection of RNA after complementary DNA synthesis on SMR biosensor, that offer simple, rapid, label-free, and real-time multiplexed detection of biomolecules near the sensor surface by using refractive index changes [[11], [12], [13]]. For clinical use, however, the sensitivity and specificity of these technologies need to be further improved.
In vivo genome editing technologies have great clinical application potentials [14,15]. Zinc-finger nucleases (ZFNs) or transcription activator-like effectors (TALEs)-based editing tools have been introduced for DNA targeting and regulation, however, these proteins need to be individually designed for DNA binding, which remains a hurdle for regulating multiple loci [14,16]. In contrast, RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR) associated Cas9 targets specific genomic loci via a single guide RNA (sgRNA), which contains a 20-bp guide sequence followed by a 3-bp protospacer adjacent motif (PAM) and recognizes target DNA through Watson-Crick base pairing [17]. The system uses an endonuclease Cas9 for nucleotide sequence based DNA targeting. Cas9 is guided by the sgRNA that specifically bind to and cleave double-stranded DNA in a site-specific manner [[18], [19], [20], [21]]. Recently, Cas9 nuclease can be used to enrich and detect the small amounts of tumor fragments in the circulating tumor DNA (ctDNA) or Zika virus [22,23]. Also, other types of Cas proteins such as Cas12a or Cas13 are used for rapid detection of nucleic acids [[24], [25], [26], [27]]. In addition to wild-type Cas9 or Cas9 orthologue proteins, catalytically inactivated Cas9 protein (dCas9) has great potential for various biological studies [28,29]. In contrast to the wild-type Cas9, the nuclease-deficient dCas9 have the ability to bind to DNA using sgRNA without cutting. Hence, dCas9 technology is a widely useful tool for in vivo and in vitro diagnostics [17,[28], [29], [30]].
In this study, we developed an improved diagnostic tool by combining a CRISPR/dCas9 and an isothermal diagnostic approach based on SMR biosensor for simultaneous nucleic acid (RNA and DNA) amplification and detection with speed as well as high sensitivity and specificity. The biosensors transduce the presence of target molecules based on binding-induced changes in the refractive index proximal to the waveguide surface [13]. In the case of combination of dCas9 and isothermal based biosensor, dCas9 can recognize the target DNA or cDNA but not cut the sequences of target. Because of the binding property of dCas9, the refractive index is changed based on the binding with target and dCas9 that could be more enhanced the sensitivity than SMR biosensor alone. On the other hand, Cas9 can recognize the target DNA or cDNA and cut the target sequences. Thus, the binding-induced changes are not affected. We demonstrated the clinical usefulness of this technology by using 3 Orientia tsutsugamushi, the causative agent of scrub typhus (ST), and 3 bunyavirus, the causative agent of severe fever with thrombocytopenia syndrome (SFTS). ST and SFTS are tick-borne infectious diseases that are common in Eastern Asia, especially in Korea, China, and Japan [28,29]. Moreover, the clinical presentations of these diseases substantially overlap; therefore, a rapid and highly sensitive detection method is greatly needed. Our method clearly distinguished between ST and SFTS within 20 min from serum samples.
2. Material and methods
2.1. Protein purification
For purification of recombinant dCas9 protein, T7 Express BL21 (DE3) E.coli were transformed with pET28a-His6-dCas9 plasmid. After culturing E.coli in Luria-Bertani (LB) broth at 30 °C, protein expression was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16 h at 16 °C. Cell pellet was collected by centrifugation at 5000g, and then lysed by sonication in lysis buffer (NaH2PO4 50 mM, NaCl 300 mM, imidazole 10 mM pH 8.0, 1 mM PMSF, 1 mM DTT, 1 mg/ml lysozyme). Soluble lysate was obtained by centrifugation at 8000g and incubated with Ni-NTA agarose beads for 1–2 h (Qiagen). Protein bound Ni-nitrilotriacetic acid (NTA) agarose beads were washed (NaH2PO4 50 mM, NaCl 300 mM, imidazole 20 mM pH 8.0) and dCas9 protein was eluted with imidazole containing buffer (NaH2PO4 50 mM, NaCl 300 mM, imidazole 250 mM pH 8.0). With 100 K Amicon centrifugal filter (Millipore), the buffer of eluted protein was exchanged, concentrated, and analyzed with 4–12% Bis-Tris gels (ThermoFisher).
2.2. In vitro cleavage assay
The PCR products (400 ng), which contained each Orientia Tsutsugamushi and bunyavirus DNA sequence, were mixed with 5.9 μl of rehydration buffer and 0.5 μl of 280 mM MgAc solution supplied in TwistAmp Basic RT kit. In 10 μl reactions, buffer mixed PCR products were incubated with 1 μg Cas9 protein and 750 ng sgRNA for 1 h at 37 °C as previous study [33]. For the positive control of the cleavage assay, same PCR products were cleaved in 1× NEBuffer 3.1 (100 mM NaCl, 50 mM Tris−HCl, 10 mM MgCl2, 100 μg/ml BSA, New England BioLabs) condition. The RNase A (4 μg) was added to samples to remove sgRNA and the final samples were analyzed with agarose gel electrophoresis.
2.3. In vitro binding assay
dsDNA templates were prepared by annealing the 5′ biotinylated target DNA strands and the non-biotinylated non-target DNA strands at a 1:1.5 molar ratio. (OT_1_F_biotin: TATAAAGATCTTGTTAAATTGCAGCGTCATGCAGGAATTAGGAAAGC, OT_1_R:GCTTTCCTAATTCCTGCATGACGCTGCAATTTAACAAGATCTTTATA, SFTS_F_biotin:AAAAATTAGCTGCCCAACAAGAAGAAGATGCAAAGAATCAAGGTGAA, SFTS_R: TTCACCTTGATTCTTTGCATCTTCTTCTTGTTGGGCAGCTAATTTTT) 10 nM dsDNA was incubated with 300 nM dCas9 and 1 μM sgRNA in cleavage buffer condition. After 20 min incubation at 37 °C, samples were resolved with 10% TBE gels using 0.5× TBE buffer supplemented with 5 mM MgCl2. Then, in vitro binding status was analyzed by using Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFisher) and Biodyne B Nylon Membrane (ThermoFisher) according to the manufacturer’s protocol.
2.4. Operation of CRISPR/dCas9-mediated biosensor and SMR biosensor alone
For simultaneous amplification and detection of target DNA or RNA using SMR biosensor alone, we used the RPA and reverse transcription (RT)-RPA solution for DNA and RNA, respectively. To prepare RPA or RT-RPA solution, 29.5 μl rehydration buffer, 15 μl of RNase inhibitor and H2O, 2.5 μl of 10 μM another primer were mixed. Then, reaction mix was added to freeze-dried enzyme. After that, 2.5 μl of 280 mM magnesium acetate (MgAc) was dissolved into the tube. After mixing, 50 μl of reaction buffer was split into five 10 μl aliquots. To begin the reactions, 5 μl of DNA or RNA extracted from blood serum of the patients and 3 μl of dCas9 RNPs (300 ng of dCas9 and 225 ng of gRNA) were added to 10 μl reaction aliquot. The volume of 18 μl mixture was added into a sensor chip with an acrylic well placed surround the microring sensor area and 10 μl mineral oil added to prevent evaporation the mixture during amplification. The chip was then placed on a thermal pad to keep a specific temperature (38 °C for DNA and 43 °C for RNA). The wavelength shift was measured every 5 min to monitor the amplification of target DNA or RNA. Relative resonant wavelength shift was calculated by the equation; ΔΔpm = (target wavelength value, pm) − (non-target wavelength value, pm).
2.5. Clinical specimens
ST and SFTS serum samples were collected from the patients in Asan Medical Center. The study protocol was approved by the Institutional Review Board of Asan Medical Center, and informed consent was obtained from all participants. SFTS was confirmed by detecting viral ribonucleic acid (RNA) by real-time RT-PCR in serum, using a DiaStar 2X OneStep RT-PCR Pre-Mix kit (SolGent, Daejeon, South Korea). A diagnosis of ST was established when we observed either a single positive result of an immunofluorescence assay (IFA; SD Bioline Tsutsugamushi Assay; Standard Diagnostics, Yongin, South Korea), or a ≥1:640 or fourfold rise of IFA titer in successive samples.
3. Results and discussion
3.1. CRISPR/dCas9-mediated biosensor as a molecular diagnostic tool
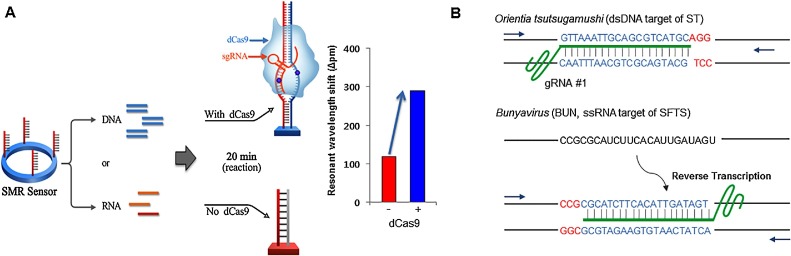
Fig. 1 A shows the design of CRISPR/dCas9-mediated biosensor that couples SMR-based isothermal nucleic acid amplification and dCas9 RNP. For simultaneous amplification and detection of nucleic acid, sequence specific primer of target was immobilized to the surface of the SMR biosensor and dCas9 RNP was in reaction chamber with single temperature for isothermal reaction with RPA. For DNA, a primer binds a recombinase enzyme to extend the DNA. For RNA, RT-RPA was used for the formation of complex of a primer and a recombinase enzyme to transcribe cDNA from RNA. The amplified nucleic acid targets were simultaneously detected by monitoring the wavelength shift. During the nucleic acid amplification process with dCas9 RNP on the SMR biosensor, the immobilized primer was hybridized with the target templates; dCas9 RNP, which allows sequence specific binding to the target nucleic acids (nearby the waveguide surface at immobilized forward primer, Supplementary Fig. S1), subsequently caused a dramatic increase as a signal enhancer in the proportion of each resonant wavelength. The SMR biosensors transduce the presence of target molecules based on binding-induced changes in the refractive index proximal to the waveguide surface. dCas9 binds to the amplified product on the surface of the sensor and increases the molecular weight of the sensor surface, thereby increasing the detection sensitivity by increasing the refractive index change. Therefore, the CRISPR/dCas9-mediated biosensor detected the pathogenic nucleic acids with high sensitivity within 20 min compared to the SMR alone (Fig. 1A).
Fig. 1.
CRISPR/dCas9-mediated biosensor. (A) Schematic of CRISPR/dCas9-mediated biosensor. SMR biosensor, silicon microring resonator sensor; dsDNA, double stranded DNA. (B) The schematics of gRNA design targeting ST (Orientia tsutsugamushi) and SFTS (Bunyavirus). The target site is highlighted in blue and the PAM sequence is shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.2. Assay optimization
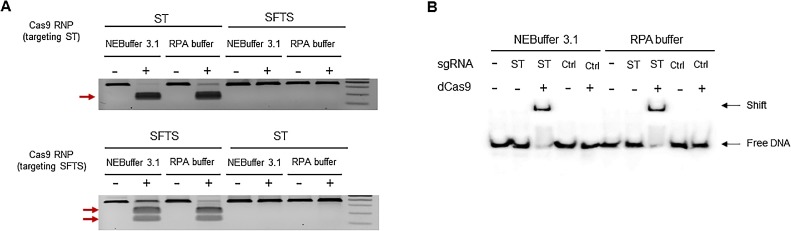
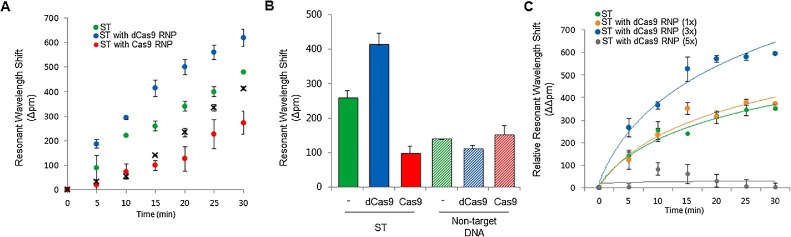
To achieve sensitive detection with dCas9 RNP on SMR biosensor, we constructed guide RNAs (gRNAs) targeting two tick-borne pathogens that have substantially overlapping clinical presentations: Orientia tsutsugamushi, the causative agent of scrub typhus (ST), and bunyavirus, the causative agent of severe fever with thrombocytopenia syndrome (SFTS) (Fig. 1B). We observed that Cas9 RNPs induced targeted DNA cleavage and dCas9 RNPs bound target DNA in RPA buffer conditions using an in vitro cleavage assay and an electrophoretic mobility shift assay (EMSA), respectively (Fig. 2 A, B). To determine whether dCas9 RNP could enhance the detection sensitivity of the SMR biosensor, we amplified DNA fragments from ST clinical samples; as a result, we observed that the signal was higher with ST + dCas9 RNPs than with ST alone or ST + Cas9 RNPs at all time points (from 5 min to 30 min) (Fig. 3 A). The relative resonant wavelength shift results of the ST with dCas9 RNPs showed that it is possible to clearly separate the positive and negative than ST alone and ST with Cas9 RNPs. In the case of ST with Cas9 RNPs, because of the cutting property of Cas9 RNPs, the result of relative resonant wavelength shift was reduced and looked like a negative. This improvement is due to the specific binding of dCas9 RNPs with the target fragment on the biosensor, and not a non-specific binding effect (Fig. 3B). Remarkably, dCas9 RNPs improved both detection sensitivity and specificity. We also optimized the concentration of dCas9 RNPs for sensitive and specific detection of pathogenic nucleic acids (Fig. 3C).
Fig. 2.
Ability of dCas9 ribonucleoprotein (RNP) in reaction buffer. (A) in vitro cleavage assay to investigate the activity of gRNAs in the RPA buffer condition. Cas9 RNP could cleave the PCR products in both the RPA buffer and the NEBuffer 3.1 condition only when gRNAs were matched to the target PCR products. (B) Electrophoretic mobility shift assay (EMSA) using dCas9 RNP and the 5′ biotinylated DNA duplexes. The target DNA duplexes were only shifted with the matched gRNAs in both the RPA buffer and the NEBuffer 3.1 condition.
Fig. 3.
Characterization of CRISPR/dCas9-mediated biosensor. (A) CRISPR/dCas9-mediated biosensor detection of ST-DNA within 30 min. ST (SMR biosensor only, green), ST with dCas9 ribonucleoprotein (RNP) (dCas9 with biosensor, blue), ST with Cas9 RNP (Cas9 with biosensor, red), and negative control (black-X). (B) Resonant wavelength shift of CRISPR/dCas9-mediated biosensor for detection of ST in 15 min. SMR biosensor alone (green), with dCas9 RNP (blue) and with Cas9 RNP (red). (C) Relative resonant wavelength shift of CRISPR/dCas9-mediated biosensor in 30 min. The colors represent the amount of the dCas9 RNP; green (ST without dCas9 RNP), orange (ST with 1× of dCas9 RNP, p > 0.1), blue (ST with 3× of dCas9 RNP, p < 0.001), and grey (ST with 5× of dCas9 RNP, p > 0.1). Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.3. Single molecule detection using CRISPR/dCas9-mediated biosensor
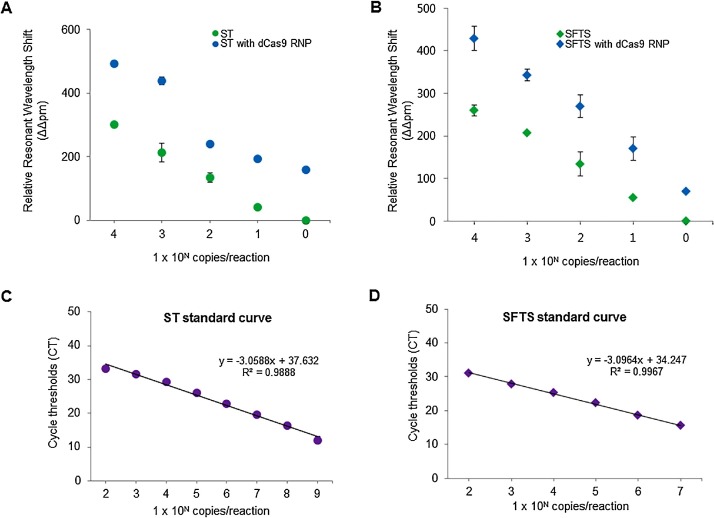
Next, we determined the detection sensitivity of CRISPR/dCas9-mediated biosensor for detection of pathogenic DNA (for ST) or RNA (for SFTS) fragments from Orientia tsutsugamushi and bunyavirus. We used a RPA- or RT-RPA based CRISPR/dCa9 mediated biosensor for double-stranded DNA and single-stranded RNA amplification, respectively. To test the detection sensitivity of ST and SFTS alone, and with dCas9 RNPs, we used serially diluted samples containing 1 × 10° to 5 × 109 copies of the ST amplicon and T7 transcribed SFTS RNA. As a result, by using dCas9 RNPs, we achieved single-molecule sensitivity for the detection of ST (0.54 aM) and SFTS (0.63 aM) within 30 min (Fig. 4 A, B). The detection sensitivity of this CRISPR/dCas9-mediated biosensor (1 copy) was superior to that of the SMR biosensor alone (∼10 copies) (Fig. 4A, B) and real-time PCRs (∼100 copies) (Fig. 4C, D).
Fig. 4.
Comparison of detection limit of CRISPR/dCas9-mediated biosensor and real-time PCRs for DNA and RNA. (A) CRISPR/dCas9-mediated biosensor can detect dsDNA of ST at concentration down to 0.54 aM (blue circle), more sensitive than biosensor alone (green circle). (B) CRISPR/dCas9-mediated biosensor detected RNA fragment of SFTS at a concentration of 0.63 aM (blue triangle), which was more sensitive than biosensor alone (green triangle). (C) Linear relationship between the concentration of target DNA and Ct value of fluorescence signal by real-time PCR. The target DNA at low concentration (<100 copies/ml) was not detected (over 40 Ct value). (D) Linear relationship between the concentration of target RNA and Ct value of fluorescence signal by real-time RT-PCR. The RNA target at low concentration (<100 copies/ml) was not detected (over 40 Ct value). Error bars indicate standard deviation from the mean, based on at least 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.4. Utility of CRISPR/dCas9-mediated biosensor in clinical samples
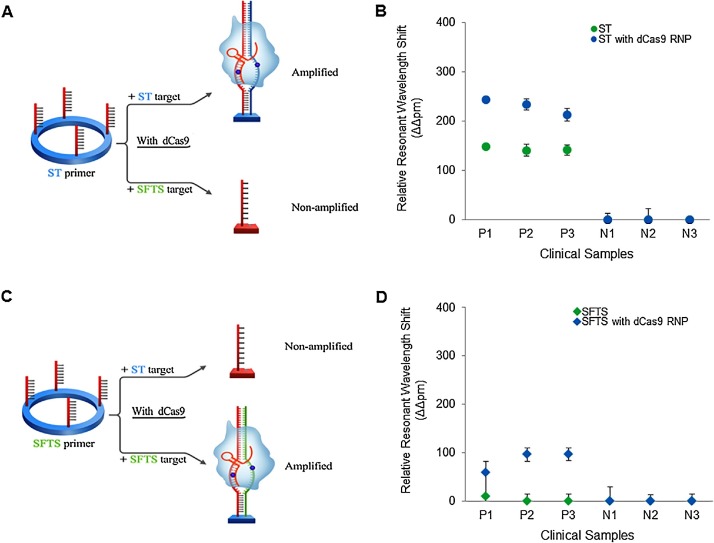
Finally, we investigated the clinical utility of this CRISPR-mediated biosensor for clinical applications that require speed, high sensitivity, and specificity, such as the diagnosis of incipient tick-borne illnesses. We used the CRISPR/dCas9-mediated biosensor to analyze six clinical samples—three from ST patients and three from SFTS patients (Fig. 5 ). When we employed the CRISPR/dCas9-mediated biosensor with ST primers, we observed elevated signals only in ST samples and not in SFTS samples (Fig. 5A). When we employed the CRISPR-mediated biosensor with SFTS primers, we detected elevated signals only in SFTS samples. Remarkably, the SFTS samples were more clearly detected by the CRISPR/dCas9-mediated biosensor than by the SMR biosensor alone (Fig. 5B). Thus, we demonstrated that the CRISPR-mediated biosensor can clearly distinguish the two pathogens from clinical samples.
Fig. 5.
Clinical utility of CRISPR/dCas9-mediated biosensor. (A) Schematic of CRISPR/dCas9-mediated biosensor for ST diagnosis in ST and SFTS samples. (B) Highly sensitive and specific detection of ST-DNA with CRISPR/dCas9-mediated biosensor (blue circle) in six clinical samples compared to that of SMR biosensor alone (green circle). P1-3, human sera from ST patients as positives; N1-3, human sera from severe fever with thrombocytopenia syndrome (SFTS) patients as negatives. (C) Schematic of CRISPR/dCas9-mediated biosensor for SFTS diagnosis in ST and SFTS samples. (D) Highly sensitive and specific detection of SFTS-RNA with CRISPR/dCas9-mediated biosensor (blue triangle) in six clinical samples compared to that of biosensor alone (green triangle). P1-3, human sera from SFTS patients as positives; N1-3, human sera from ST patients as negatives. Error bars indicate standard deviation from the mean, based on at least three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4. Conclusion
We developed a CRISPR/dCas9-mediated biosensor that combines a catalytically inactive dCas9 and SMR biosensor based isothermal nucleic acid amplification for rapid and high sensitive detection of pathogens in clinical settings. Using CRISPR/dCas9-mediated biosensor, we achieved single molecule sensitivity for the detection of ST (0.54 aM) and SFTS (0.63 aM). Also, our method clearly distinguished between ST and SFTS within 20 min from serum samples.
We addressed several features for clinical use of this assay. First, CRISPR/dCas9-mediated biosensor enhances detection sensitivity through triple-targeting (primer, target strand, and dCas9). dCas9 shows higher sensitivity and specificity than the conventional method because it requires accurate target recognition of 20 nt of gRNA (Supplementary Table S2). Importantly, the triple-targeting method provides a significant advancement in terms of detection sensitivity from our previous SMR biosensor approach that utilized primer to target strand only [11,13]. By using triple-targeting, we were able to detect a single copy target, which translates into 10 times higher sensitivity than that of SMR biosensor alone (10 copies) and 100 times more sensitive than that of real-time (RT)-PCR methods (100 copies). Second, the readout system of CRISPR/dCas9-mediated biosensor differs from other CRISPR based diagnostic assays (i.e., fluorescence dye) in fundamental ways [[24], [25], [26], [27]]. SMR biosensor based isothermal nucleic acid amplification assay can be simultaneously amplified and detect the target without labeling. In addition, CRISPR/dCas9 enhanced the refractive index change on the sensor surface. Therefore, the readout of sample using our assay can be carried out within 20 min. Third, the results of false-positive or false-negative are minimized by the use of sgRNA, primer, and dCas9. We have also shown that this assay is able to clearly distinguish the two pathogens in clinical samples. Our data on the clinical sensitivity and specificity highlights the utility and practicality of CRISPR/dCas9-mediated biosensor. Fourth, Gootenberg et al. developed a CRISPR/Cas13a (previously known as C2c2)-based diagnostic assay [24,25]. Because Cas13a is a programmable RNA-guided ribonuclease, Cas13a-based diagnosis is an RNA-based detection method that exploits the collateral effect of Cas13a. Our complementary approach using CRISPR/dCas9 is a DNA-based detection method that is able to detect nucleic acids during isothermal nucleic acid amplification without in vitro RNA transcription.
Finally, our CRISPR/dCas9-mediated biosensor method is being further improved for convenient operation based on the ASSURED criteria (Affordable, Sensitivity, Specificity, User-friendly, Rapid, Equipment-free, Deliverable) in clinical settings [30]. A microfluidic platform is being developed for sample processing that uses non-chaotropic reagent-based thin film techniques. The platform is devised to allow simple, low-cost, rapid, and high-throughput nucleic acids (DNA, RNA) extraction from human body fluids [31,32]. We believe that the final integration of sample preparation and CRISPR/dCas9-mediated biosensors into a single cartridge will significantly facilitate emerging pathogen diagnosis for making timely treatment decision for emerging infectious diseases.
Acknowledgements
This study was supported by the Ministry of Science, ICT and Future Planning (MSIP) and the Ministry of Education through the National Research Foundation of Korea (NRF) (2017R1A2B4005288, 2016R1D1A1A02937096).
Biographies
Bonhan Koo received the M.S degree in University of Ulsan College of Medicine in 2018. He is a Ph.D student at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. His research is focused on development of molecular diagnostic platform based on optical biophotonics for detection of disease related biomarkers.
Da-Eun Kim is a Ph.D student at Seoul National University, Seoul, Korea. Her research is now focused on study of CRISPR/Cas9 function in clinical applications.
Jiyeon Kweon received the Ph.D degree in Seoul National University, Seoul, Korea. She is a postdoctoral fellow at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. Her research is now focused on study of CRISPR/Cas9 function in clinical applications.
Choong Eun Jin received the M.S degree in SangMyung University in 2016. She is a Ph.D student at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. Her research is focused on development of molecular diagnostic platform based on optical biophotonics for detection of disease related biomarkers.
Sung-Han Kim received the M.D and Ph.D. degree in Seoul National University School of Medicine, Seoul, Korea. He is an associate professor at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. His research is now focused on clinical application using new technologies in infectious diseases.
Yongsub Kim received the Ph.D degree in Seoul National University, Seoul, Korea. He is an assistant professor at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. His research is now focused on study of CRISPR/Cas9 function in clinical applications.
Yong Shin received the Ph.D. degree in Max Planck Institute of Experimental Medicine and Georg-August-University Goettingen, Germany in 2008. He is an associate professor at University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. His research is now focused on development of molecular diagnostic platform based on optical biophotonics for detection of disease related biomarkers.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2018.06.069.
Contributor Information
Yongsub Kim, Email: yongsub1.kim@gmail.com.
Yong Shin, Email: shinyongno1@gmail.com.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zhang B., Pinsky B.A., Ananta J.S., Zhao S., Arulkumar S., Wan H. Diagnosis of Zika virus infection on a nanotechnology platform. Nat. Med. 2017;23:548–550. doi: 10.1038/nm.4302. [DOI] [PubMed] [Google Scholar]
- 2.Broadhurst M.J., Brooks T.J., Pollock N.R. Diagnosis of Ebola virus disease: past, present, and future. Clin. Microbiol. Rev. 2016;29:773–793. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guery B., Poissy J., el Mansouf L., Séjourné C., Ettahar N., Lemaire X. Clinical features and viral diagnosis of two cases of infection with middle east respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson M.L. Malaria rapid diagnostic tests. Clin. Infect. Dis. 2012;54:1637–1641. doi: 10.1093/cid/cis228. [DOI] [PubMed] [Google Scholar]
- 5.Boelaert M., Bhattacharya S., Chappuis F., El Safi S.H., Hailu A., Mondal D. Evaluation of rapid diagnostic tests: visceral leishmaniasis. Nat. Rev. Microbiol. 2007;5:S20–S39. [Google Scholar]
- 6.Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J. Genetic and serologic properties of Zika virus associated with an epidemic, Yap state, micronesi, 2007. Emerg. Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos Rde M., Cirne-Santos C., Meira G.L., Santos L.L., de Meneses M.D., Friedrich J. Prolonged detection of Zika virus RNA in urine samples during the ongoing Zika virus epidemic in Brazil. J. Clin. Virol. 2016;77:69–70. doi: 10.1016/j.jcv.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Tappe D., Rissland J., Gabriel M., Emmerich P., Gunther S., Held G. First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Euro Surveill. 2014;19:20685. doi: 10.2807/1560-7917.es2014.19.4.20685. [DOI] [PubMed] [Google Scholar]
- 9.Baker M. New-wave diagnostics. Nat. Biotechnol. 2006;24:931–938. doi: 10.1038/nbt0806-931. [DOI] [PubMed] [Google Scholar]
- 10.Yoo S.M., Lee S.Y. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016;34:7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Shin Y., Perera A.P., Kim K.W., Park M.K. Real-time, label-free isothermal solid-phase amplification/detection (ISAD) device for rapid detection of genetic alteration in cancers. Lab Chip. 2013;13:2106–2114. doi: 10.1039/c3lc50129a. [DOI] [PubMed] [Google Scholar]
- 12.Shin Y., Perera A.P., Tang W.Y., Fu D.L., Liu Q., Sheng J.K. A rapid amplification/detection assay for analysis of mycobacterium tuberculosis using an isothermal and silicon bio-photonic sensor complex. Biosens. Bioelectron. 2015;68:390–396. doi: 10.1016/j.bios.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Koo B., Jin C.E., Lee T.Y., Lee J.H., Park M.K., Sung H. An isothermal, label-free, and rapid one-step RNA amplification/detection assay for diagnosis of respiratory viral infections. Biosens. Bioelectron. 2017;90:187–194. doi: 10.1016/j.bios.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 15.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaj T., Gersbach C.A., Barbas C.F. ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S. CRISRP provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 20.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.H., Yu J., Hwang G.H., Kim S. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene. 2017;36:6823–6829. doi: 10.1038/onc.2017.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W. Rapid, Low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 24.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R. Two distinct RNase activities of CRISRP-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J.S., Ma E., Harrington L.B., Da Costa M. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder J.C., Bateson M.M., Lavin M., Young M.J. Use of cellular CRISPR (Clusters of Regularly Interspaced Short Palindromic Repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl. Environ. Microbiol. 2010;76:7251–7258. doi: 10.1128/AEM.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Method. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balboa D., Weltner J., Eurola S., Trokovic R., Wartiovaara K., Otonkoski T. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Rep. 2015;5:448–458. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seong S.Y., Choi M.S., Kim I.S. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 2001;3:11–21. doi: 10.1016/s1286-4579(00)01352-6. [DOI] [PubMed] [Google Scholar]
- 32.Yun Y., Heo S.T., Kim G., Hewson R., Kim H., Park D. Phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in south korea and migratory bird routes between china, south korea, and japan. Am. J. Trop. Med. Hyg. 2015;93:468–474. doi: 10.4269/ajtmh.15-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Bae S., Park J., Kim E. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.