Figure 1.
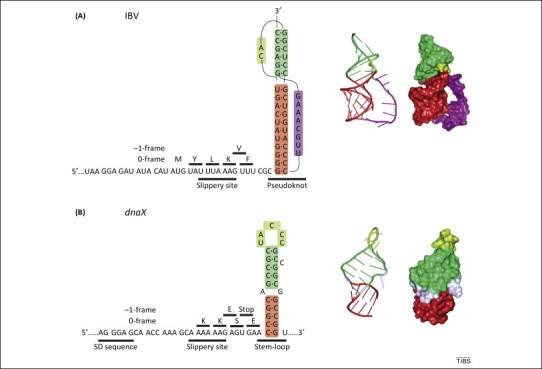
Frameshifting stimulatory signals on mRNA. (A) Left panel, schematic of the modified frameshifting sequence of the avian infectious bronchitis virus (IBV) 1a/1b mRNA [27]. The original IBV slippery sequence (U UUA AAC) was changed to ensure maximum frameshifting efficiency in Escherichia coli[37]. The amino acids incorporated upon translation in the 0- or −1-frame are indicated above the mRNA sequence. Middle and right panels, structure and surface representation of the frameshifting pseudoknot. (B) Left panel, schematic of the programmed −1 ribosomal frameshifting (−1PRF) regulatory elements in dnaX from E. coli. In addition to the slippery sequence and stimulatory stem-loop, frameshifting on dnaX is modulated by a Shine–Dalgarno (SD)-like sequence upstream of the slippery site [22]. Middle and right panels, structure and surface representation of the frameshifting stem-loop. Images were prepared using PyMOL (http://www.pymol.org/).
