Figure 2.
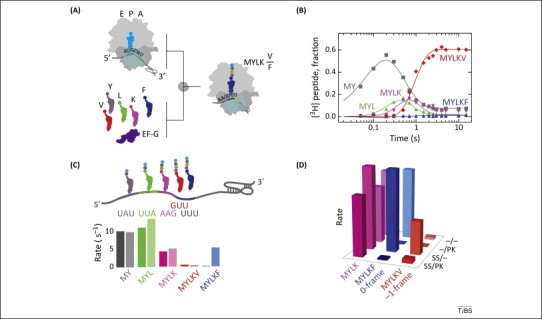
Codon walk over the frameshifting sequence of avian infectious bronchitis virus (IBV). (A) Schematic of the experiments. Purified translation components are rapidly mixed in a quench-flow device. After the desired time the reactions are stopped by rapid mixing with a strong base (quencher) and the peptide products are analyzed by HPLC, measuring [3H]Met radioactivity. (B) Example of a time-course of peptide synthesis on a frameshifting mRNA. Monitored peptides are fMetTyr (MY), fMetTyrLeu (MYL), fMetTyrLeuLys (MYLK), fMetTyrLeuLysPhe (MYLKF), and fMetTyrLeuLysVal (MYLKV). The time-courses are used to calculate the rates and efficiencies of insertion into peptide of each amino acid in 0- and −1-frame. (C) Comparison of the amino acid incorporation rates (s−1) for frameshifting (dark colors) and non-frameshifting (light colors) mRNAs. The frameshifting mRNA contained both a slippery sequence and pseudoknot, the non-frameshifting mRNA had no stimulatory elements for programmed −1 ribosomal frameshifting (−1PRF). (D) Contributions of the regulatory elements in the mRNA. SS and PK indicate the presence of a slippery site or a pseudoknot, respectively, in the mRNA [27].
