Figure 3.
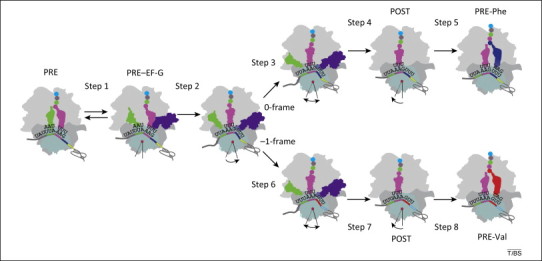
Kinetic model of programmed −1 ribosomal frameshifting (−1PRF). Frameshifting occurs during translocation of the two tRNAs bound to the slippery sequence (tRNALeu in the P site and MYLK-tRNALys in the A site). Recruitment of EF-G (step 1) to the pre-translocation (PRE) complex facilitates rapid tRNA movement (step 2) into a state where translocation on the 50S subunit is completed; however, the following steps are inhibited by the presence of the pseudoknot. tRNALeu moves on the 50S subunit while the distance to the 30S subunit is not changed (steps 3 and 6 in 0-frame and −1-frame, respectively). Then, tRNALeu and the 30S subunit move apart (steps 4 and 7). Steps 3 and 4 are particularly slow for the tRNA that remains in 0-frame, which limits the decoding rate in the 0-frame by Phe-tRNAPhe (step 5). By contrast, tRNALeu movement on those ribosomes that shift to the −1-frame is faster (step 6), followed by dissociation of tRNALeu from the 30S subunit, 30S head rotation, dissociation of EF-G (step 7), and binding of Val-tRNAVal (step 8). Reproduced, with permission, from [27].
