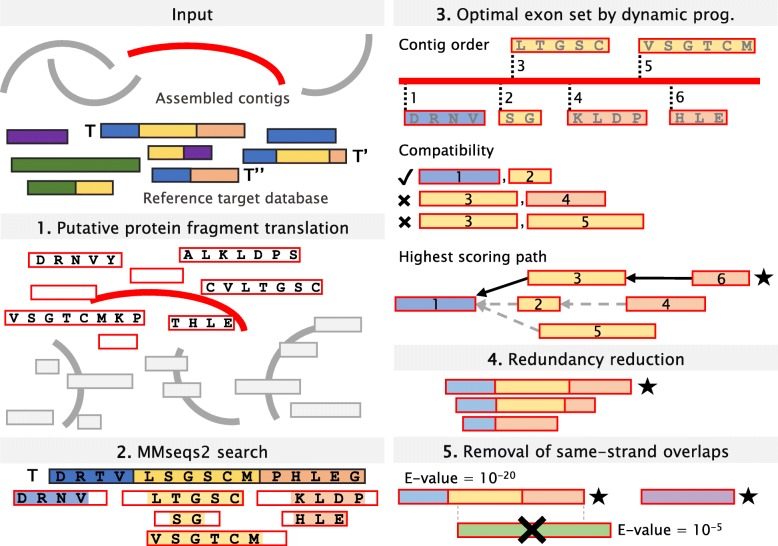
Fig. 1.
MetaEuk algorithm. Input to MetaEuk are assembled metagenomic contigs and a reference database of protein sequences. 1 Six-frame translation of all putative protein-coding fragments from each contig. 2 Fragments on the same contig and strand that hit the same reference protein T are examined together. 3 Putative exons are identified and ordered according to their start position on the contig. The highest score and path (denoted with a star) of a set of compatible putative exons is computed by dynamic programming, in which individual scores of the putative exons are summed and unmatched amino acids are penalized. 4 Redundancies among gene calls due to homologous targets (T, T’, and “T”) are reduced and a representative prediction (denoted with a star) is retained. 5 Contradicting predictions of overlapping genes on the same strand are resolved by excluding the prediction with the higher E-value