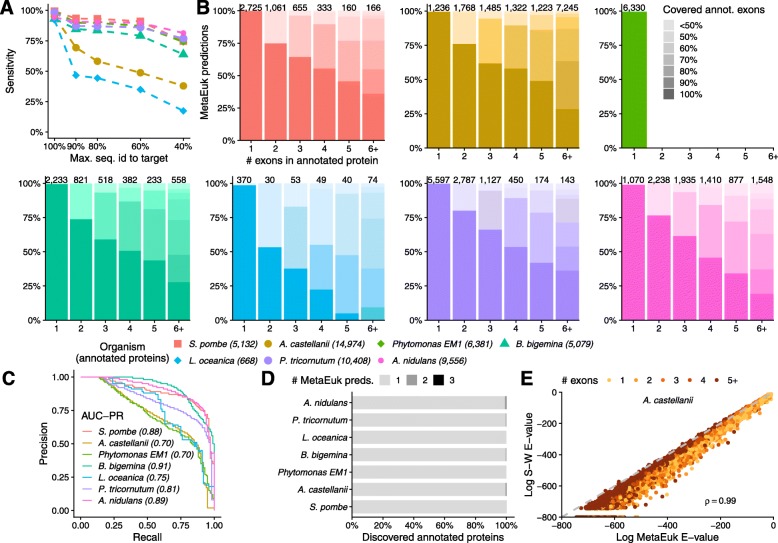
Fig. 2.
MetaEuk evaluation on benchmark. MetaEuk predictions were mapped to annotated proteins. a Conditions of increasing evolutionary divergence were simulated by excluding gene calls based on their sequence identity to their target. Sensitivity is the fraction of annotated proteins from the query genome to which a MetaEuk prediction was mapped. b Fraction of exons covered by MetaEuk (color saturation). The number of MetaEuk predictions is indicated on top of each bar. c In an annotation-dependent precision estimation, MetaEuk predictions that mapped to an annotated protein were considered as “true” and the rest as “false.” These sets of predictions are well separated by their E-values, as indicated by the high AUC-PR values. d Fraction of annotated protein-coding genes that were split by MetaEuk into two (dark grey) or three (black) different predictions. e Comparison of the E-values computed by MetaEuk and by the Smith-Waterman algorithm for A. castellani proteins. The Spearman rho values indicate high correlation for A. castellani and for the other organisms (Supp. Figure S3A)