Figure 8.
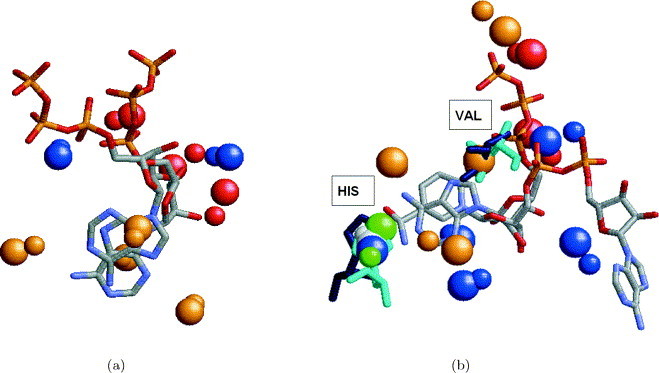
Alignment between the ligands induced by the alignment of the binding sites. (a) Alignment between the ATP ligands of hexamerization domain of N-ethylmaleimide sensitive factor (1nsf) and cAMP-dependent protein kinase (1atp) obtained by the alignment of the corresponding binding sites. The colored spheres represent the centers of interaction recognized by the program. The coloring of the spheres is as in Figure 2. The spheres of 1nsf are smaller. It can be seen that the solution provides a good alignment between the ribose parts of the two ATP molecules at the expense of the alignment between the adenine moieties. (b) Alignment between the NAD and ATP ligands of lactate dehydrogenase (9ldt) and hypothetical protein MJ0577 (1mjh) obtained by the alignment of the corresponding binding sites. The solution provides a good alignment between the ribose parts of the NAD and ATP ligand molecules. However, the adenine ring of ATP is aligned to a nicotinamide ring of NAD, and not to its adenine ring as expected. As can be seen, the resulting alignment provides a superimposition of 13 functional groups, that are depicted as balls. Moreover, it provides an alignment of two conserved residues His40 and Val142 of 1mjh (colored cyan) and His195, Val32 of 9ldt (colored blue) that are located in similar spatial locations.
