Figure 3.
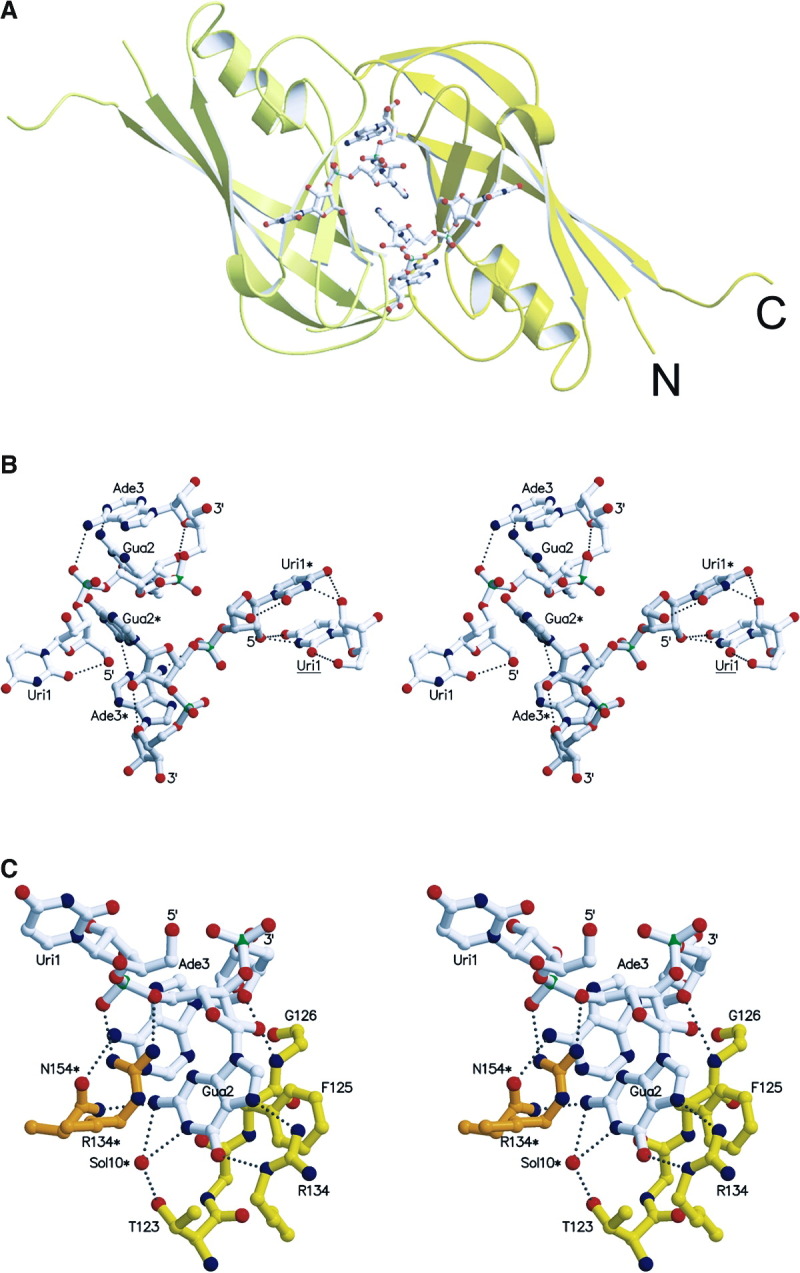
RNA Conformation and RNA Protein Interactions
(A) Two RNA molecules bind at the molecular interface generated by the dimer-dimer formation. The two protomers are shown in yellow and light yellow and the triribonucleotide is drawn as an all-atom model.
(B) Stereo view of two complete symmetry-related triribonucleotides containing the sequence 5′-U-G-A-3′, bound at the dimer-dimer interface. The two molecules are in white and in light gray with labels containing one asterisk. In addition, the right panel includes also a uridine (label underlined) from a neighboring RNA molecule, interacting specifically with Uri1* (for clarity, only Uri1 of the triribonucleotide is shown). The hydrogen bonding distances within the triribonucleotides are indicated as dashed lines. Note the parallel base pair stacking by the central guanine (Gua2 and Gua2*) as well as the parallel orientation of two symmetry-related uracil bases (Uri1* and Uri1). Both riboses from the uridine and the guanine are in the C2′-endo conformation while the guanine ribose is in C3′-endo [29].
(C) Close-up stereo view of protein-RNA interactions, shown for a single RNA molecule; van der Waals contacts are not shown with the exception of the parallel base pair stacking of F125. Note that most of the interactions are contributed by the guanosine phoshate. Hydrogen bonding distances are indicated by dashed lines and contributions of symmetry-related residues are marked with an asterisk (N154*).
