Fig. 1.
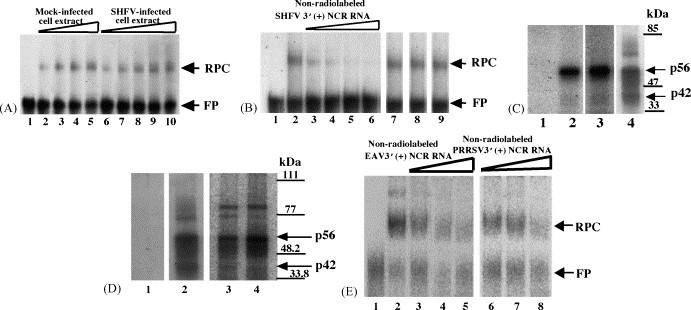
Analysis of the interaction between proteins in MA104 cell extracts and 32P-labeled arterivirus 3′(+)NCR RNAs. (A) Gel mobility shift assay. Radiolabeled SHFV 3′(+)NCR RNA was incubated with an S100 cytoplasmic extract from either SHFV-infected or mock-infected MA104 cells. The RPCs were resolved on a 10% non-denaturing polyacrylamide gel and visualized by autoradiography. (Lane 1) free probe; (lanes 2–5) increasing amounts of mock-infected MA104 S100 cytoplasmic extract (100, 200, 300, and 400 ng); (lanes 6–10) increasing amounts of SHFV-infected MA104 S100 cytoplasmic extract (100, 200, 300, 400, and 500 ng). The locations of the RNA–protein complex and free probe are indicated by arrows. (B) Competition gel mobility shift assay. Different amounts of non-radiolabeled competitor RNAs were incubated with an MA104 S100 cytoplasmic extract before addition of the 32P-labeled SHFV 3′(+)NCR RNA. The RPCs were resolved on a 10% non-denaturing polyacrylamide gel and visualized by autoradiography. (Lane 1) free probe; (lane 2) no competitor; (lanes 3–6) increasing amounts of non-radiolabeled SHFV 3′(+)NCR RNA (5-, 10-, 20-, and 30-fold molar excess); (lane 7) 250-fold molar excess of yeast tRNA; (lane 8) 150-fold molar excess of WNV 3′(+)SL RNA; (lane 9) 250-fold molar excess of poly(I)–poly(C). The locations of the RNA–protein complex and free probe are indicated by arrows. (C) UV-induced cross-linking assay. MA104 S100 cytoplasmic extracts were incubated with radiolabeled SHFV 3′(+)NCR RNA and then were exposed to UV-irradiation. The unprotected RNA was digested with RNase A and the cross-linked proteins were resolved by 10% SDS–PAGE and visualized by autoradiography. (Lane 1) free probe; (lane 2) mock-infected MA104 S100 cytoplasmic extract (1 μg) and poly(I)–(C) (1 μg); (lane 3) SHFV-infected MA104 S100 cytoplasmic extract (1 μg) and poly(I)–(C) (1 μg); (lane 4) mock-infected MA104 S100 cytoplasmic extract (1 μg) and poly(I)–(C) (600 ng). Standard protein markers are indicated by lines and the positions of p56 and p42 are indicated by arrows. (D) UV-induced cross-linking assay. MA104 S100 cytoplasmic extracts and different arterivirus 32P-labeled RNA probes were cross-linked by UV-irradiation in the presence of 600 ng of poly(I)–(C). (Lane 1) free probe; (lane 2) SHFV 3′(+)NCR RNA; (lane 3) EAV 3′(+)NCR RNA; (lane 4) PRRSV 3′(+)NCR RNA. The gels shown in lanes 1 and 2 were analyzed by autoradiography and the gel shown in lanes 3 and 4 was analyzed using the FUJI Bio Imaging Analyzer. The positions of protein standard markers are indicated by lines on the right. The positions of the p56 and p42 bands are indicated by arrows. (E) Competition gel mobility shift assay. MA104 S100 cytoplasmic extracts were incubated with different amounts of non-radiolabeled arterivirus RNAs before addition of the 32P-labeled SHFV 3′(+)NCR RNA. (Lane 1) free probe; (lane 2) no competitor; (lanes 3–5) increasing amounts of unlabeled EAV 3′(+)NCR RNA (25-, 50-, and 75-fold molar excess); (lanes 6–8) increasing amounts of unlabeled PRRSV 3′(+)NCR RNA (25-, 50-, and 75-fold molar excess). The gels were analyzed using the FUJI Bio Imaging Analyzer. The locations of the RNA–protein complex and free probe are indicated by arrows.
