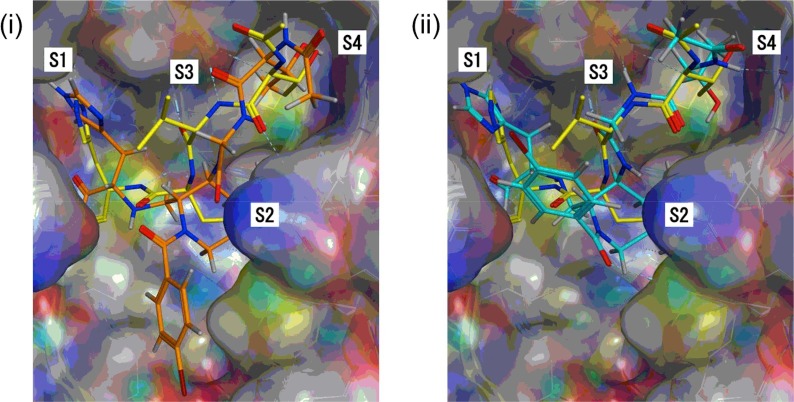
Fig. 3.
Docking model of 26 with R188I SARS 3CLpro: (i) interaction mode with GBVI/WSA dG score, −11.0665 kcal/mol: (ii) interaction mode with GBVI/WSA dG score, −10.9356 kcal/mol. Each docking model of 26 with R188I SARS 3CLpro was constructed by using an X-ray crystal structure of a complex of SARS 3CLpro and inhibitor 2 (PDB 4TWW) as a template. The possible binding mode was obtained by a docking simulation of inhibitor 26 and SARS 3CLpro using an automated template-guided docking protocol with the Amber10:EHT force field in the Molecular Operating Environment (MOE) 2018.01 software package (Chemical Computing Group Inc., Montreal, Quebec, Canada).