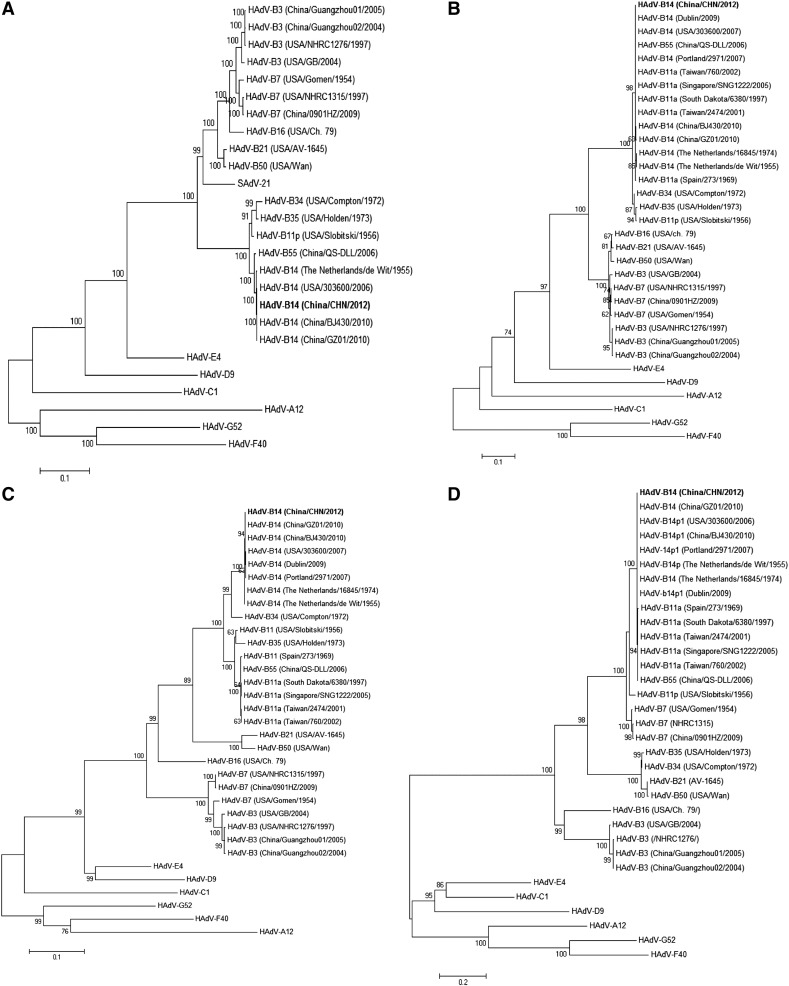
Fig. 2.
Phylogenetic analysis.
Maximum-likelihood phylogenetic trees were estimated using full-length (A) genomic sequences, (B) E1A gene sequences, (C) hexon gene sequences, and (D) fiber gene sequences of AdVs. The GenBank accession IDs and details of the full-length genomic sequences and E1A, hexon, and fiber genes sequences used in phylogenetic analysis are given in Table 1 and Table S1 respectively. Reference sequences representing different AdV genomes together with the newly sequenced HAdV-B14 CHN (bold format) were included in each tree construction dataset. Isolates were named using the following format: AdV type (sampling country/isolate name/isolation year). The percentage of trees in which the associated taxa clustered together is shown next to the branches. The reliability of the tree was assessed by bootstrap analysis with 1000 replications.