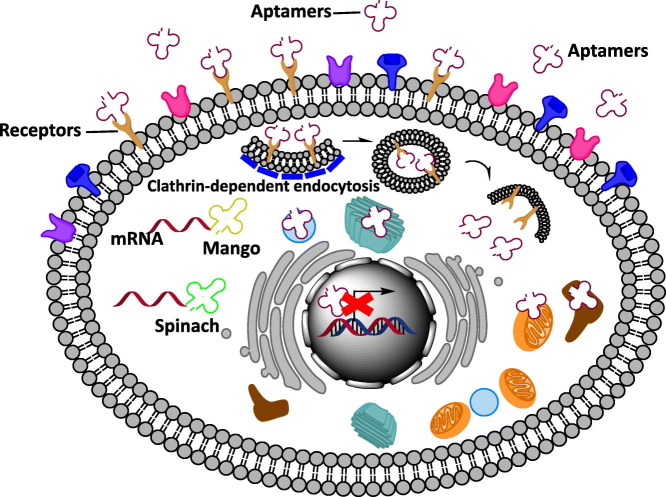
Abstract
The structural flexibility and small size of aptamers enable precise recognition of cellular elements for imaging and therapeutic applications. The process by which aptamers are taken into cells depends on their targets but is typically clathrin-mediated endocytosis or macropinocytosis. After internalization, most aptamers are transported to endosomes, lysosomes, endoplasmic reticulum, Golgi apparatus, and occasionally mitochondria and autophagosomes. Intracellular aptamers, or “intramers,” have versatile functions ranging from intracellular RNA imaging, gene regulation, and therapeutics to allosteric modulation, which we discuss in this review. Immune responses to therapeutic aptamers and the effects of G-quadruplex structure on aptamer function are also discussed.
Abbreviations: SELEX, systematic evolution of ligands by exponential enrichment; AML, amiloride; CPZ, chlorpromazine; GEN, genistein; LAMP-1, lysosomal-associated membrane protein 1; ER, endoplasmic reticulum; QD, quantum dots; NP, nanoparticles; TEM, transmission electron microscopy; PRMT5, protein arginine methyltransferase 5; DFHBI, 3,5-difluoro-4-hydroxybenzylidene imidazolinone; TO, thiazole orange; DFHO, 3,5-difluoro-4-hydroxynenzylidene imidazolinone-2-oxime; GFP, green fluorescence protein; E.coli, Escherichia coli; cdiA, cyclic di-AMP; ADP, adenosine di-phosphate; SAM, S-adenosylmethionine; GTP, guanosine 5-triphosphate; C. elegans, Caenorhabditis elegans; EGFP, enhanced green fluorescence protein; snRNA, small nuclear RNA; snRNP, small nuclear protein; apta-FRET, aptamer-based Förster resonance energy transfer; TF, transcription factor; TetR, tetracycline repressor; RNAP, RNA polymerase; RAPs, RNA polymerase binding aptamers; iRAPs, inhibitory RNA polymerase binding aptamers; UTR, Untranslated region; Hsp70, Heat shock protein 70; FOXM1, forkhead box M1; DBD, DNA binding domain; HSP, heat shock factor; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; RUNX1, runt-related transcription factor 1; RHD, runt-homology domain; USP14, ubiquitin carboxyl-terminal hydrolase 14; CBFβ, core-binding factor β protein; GPCR, G-protein-coupled receptors; HIV, human immunodeficiency virus; AD, Alzheimer's disease; TGF-β1, transforming growth factor beta 1; CSC, cancer stem cell; HER2, human epidermal growth factor receptor 2; EpCAM, epithelial cell adhesion molecule; MRP1, multidrug resistance-associated protein 1; eIF4A, eukaryotic initiation factor-4A; ATP, adenosine triphosphate; β2AR, β2-adrenoceptor; Sf9, Spodoptera frugiperda 9; cJun, protein that in humans is encoded by the JUN gene; cFos, proto-oncogene that is the human homolog of the retroviral oncogene v-fos; AP-1, activator protein-1; dsRBM, double-stranded RNA binding domain
Keywords: Aptamer, Endocytosis, Imaging, Gene regulation, Intramers, Allosteric modulation, G-quadruplex
Graphical abstract
1. Introduction
Aptamers are folded nucleic acid molecules that bind to specific targets, mimicking antibodies. Some aptamers exist naturally as ligand-binding elements, but most are generated in vitro and tailored to specific purposes. Aptamer's great advantages; structural flexibility, stability, and high affinity and specificity, and its comparably smaller size (~3 nm) than antibodies (10–15 nm) [1], allow to bind more dense cellular epitopes in living cells where the epitopes are natively folded [2]. It demonstrates that the true value of aptamers lies in their superior target recognition. As the aptamers show the better structure recognition on the intracellular epitopes, they have quickly emerged as intracellular targeting agents and intracellular imaging tools. In this review, we discuss from the uptake mechanisms, subcellular localization, and intracellular imaging to therapeutic applications of aptamers in human diseases. We also highlight specific examples of conformational selectivity of aptamers as an emerging new field, along with the future strategies to facilitate the translation of intracellular aptamers.
2. Mechanisms of internalization of aptamers
The aptamers have been developed against the cell surface receptor for targeted delivery. However, so far, few studies have investigated the endocytic mechanism of these aptamers. For the possible endocytic mechanisms, there are numerous well defined pathways by which aptamers can be internalized from the surface of cell membranes: clathrin- and caveolae-mediated endocytosis, macropinocytosis, and phagocytosis [[3], [4], [5]]. But, the uptake of aptamers mainly depends on the function of their target receptors. Based on the published studies, the mechanism of internalization of aptamers can be divided into clathrin-dependent or -independent.
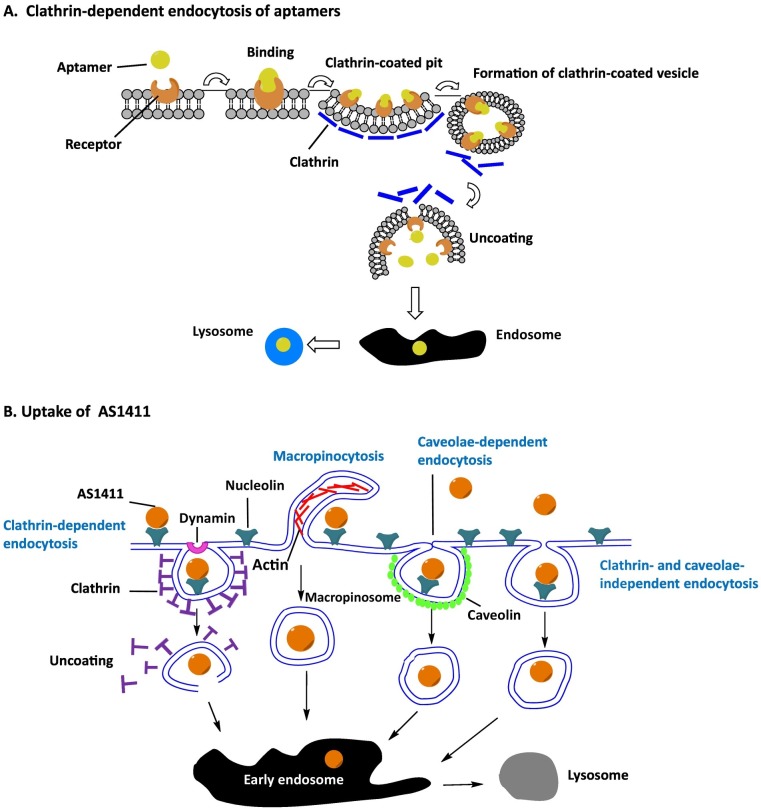
To investigate the clathrin-dependent pathway of aptamers, the colocalization study was mainly used with fluorescently labeled transferrin, because the endocytic pathway of transferrin receptor was well characterized via clathrin mediated endocytosis [6]. The representative aptamers that internalized via clathrin-dependent endocytosis were the Burkett's lymphoma cell-specific DNA aptamers and anti-protein tyrosine kinase 7 aptamers [7, 8] (Fig. 1A). Their clathrin-dependent endocytosis was confirmed with fluorescently labeled transferrin.
Fig. 1.
Mechanisms of endocytosis. (A) Clathrin-dependent endocytosis of aptamers. The binding of the aptamer and receptor initiate formation of the clathrin-coated pit, followed by clathrin-coated vesicle budding. Once detached from the membrane, the clathrin coat is disassembled. Clathrin-dependent endocytosis ends in fusion with endosomes and lysosomes. (B) Uptake mechanism of AS1411. The endocytosis of AS1411 involves multiple pathways. Although macropinocytosis is the predominant endocytosis mechanism of AS1411, clathrin- and caveolae-dependent and clathrin- and caveolae-independent pathways are all involved in uptake of AS1411.
To investigate the clathrin-independent endocytic pathway of aptamers, the inhibitors of internalization were used in most of studies. For the example of clathrin-independent pathway, AS1411 aptamers were well characterized. The G-quadruplex nucleolin DNA aptamers bound to nucleolin which was selectively expressed on the cancer plasma membrane and were internalized into cells [9, 10]. To elucidate the endocytic mechanism of AS1411, DU145 prostate cancer cells were pretreated with cytochalasin D (an actin polymerization inhibitor) and dynasore (a dynamin inhibitor). Because clathrin-mediated endocytosis depends on actin and dynamin function, whereas macropinocytosis and phagocytosis do not. Following this pretreatment, the AS1411 uptake was slightly decreased, suggesting that AS1411 was not internalized via clathrin-mediated endocytosis [11]. But, interestingly, pretreatment with amiloride (AML, a macropinocytosis inhibitor) significantly reduced the uptake of AS1411 in cancer cells. The colocalization of AS1411 with the macropinocytic marker (dextran) was confirmed again by confocal microscopy, showing the consistent results of chemical inhibitors. Therefore, AS1411 seems to be predominantly internalized into cancer cells via macropinocytosis [11].
In separate another study, the endocytic pathway of AS1411 was investigated in different PC3 prostate cancer cells. The PC3 prostate cancer cells were pretreated with AML, chlorpromazine (CPZ, a clathrin-mediated endocytosis inhibitor), and genistein (GEN, a caveolae- and lipid raft-mediated endocytosis inhibitor) [12]. Uptake of AS1411 was significantly inhibited by both AML and CPZ, but neither completely prevented aptamer internalization. It suggests that a mix of clathrin-mediated endocytosis and micropinocytosis are used for the endocytosis of AS1411 [12]. On the contrary, GEN had no effect on AS1411 uptake, demonstrating that caveolae- and lipid raft-mediated endocytosis was not involved in endocytosis of AS1411 [12]. Taken together of these two studies, it indicates that the internalization of the nucleolin aptamer AS1411 does not follow the standard pathway and also suggest that the uptake mechanisms may vary on the type of cancer cells (Fig. 1B).
3. Subcellular localization of internalized aptamers
3.1. The fate of internalized aptamers in the cytosol
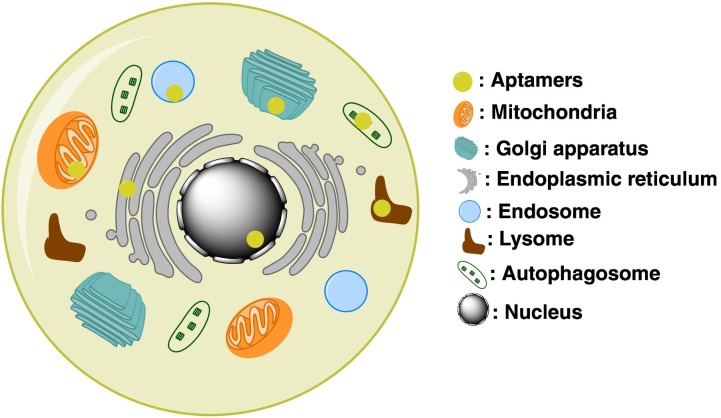
After receptor-mediated endocytosis, the aptamer-receptor complexes are fused with early endosomes and either sorted for recycling back to the plasma membrane or delivered to late endosomes and lysosomes for degradation [4]. To know the fate of internalized aptamers in cells, the subcellular distribution of aptamers has been investigated in various in vitro systems with fluorescence confocal microscopy and transmission electron microscopy (TEM) using antibodies or nanoparticles.
With the fluorescence confocal microscopy, the subcellular distribution of internalized aptamers was characterized in four studies. The colocalization of cell type specific-DNA aptamers with LAMP-1, a late endosomal and lysosomal marker, was confirmed in renal proximal tubule RPTEC/TERT1 cells by in situ fluorescence hybridization [13]. Glioma-specific DNA aptamers bound Ku 70 and Ku 80 were found to colocalize in lysosomes, endoplasmic reticulum (ER), and Golgi apparatus with organelle-specific fluorescent dyes on live cell images by confocal microscopy [14]. The PrPc DNA aptamers were conjugated with fluorescent quantum dots (QDs), which are fluorescent nanoparticles to track the dynamics of intracellular imaging. The PrPc-aptamer-QD complexes were colocalized in lysosome, ER, and the Golgi apparatus in immunofluorescence assays [15]. The RNA aptamers against human papillomavirus 16-E7 have been found in endosomes, lysosomes, autophagosomes, and ER with immunostaining by fluorescence confocal microscopy [16].
The conjugation with nanoparticles (NPs) to aptamers was used to directly observe the subcellular localization of aptamers by optical microscopy or TEM without any fluorescence tags. The nucleolin DNA aptamer AS1411 was conjugated with NPs and its subcellular location was determined by TEM in C6 brain glial tumor cells and bEnd.3 brain endothelial cells [17]. The AS1411 aptamer was mainly colocalized in endosomes and mitochondria in both C6 and bEnd.3 cells. But, the localization of AS1411 aptamers in lysosomes and the Golgi apparatus observed only in C6 cells but not bEnd.3 cells [17]. It suggests that the subcellular localization of aptamer can be varied depending on the type of cells. The anti-transcription factor activator protein-1 (AP-1) DNA aptamers (5ECdsAP1) were tagged with superparamagnetic iron oxide NPs or gold NPs [18]. The TEM study showed that the 5ECdsAP1 aptamers were localized in only ER, but not other cellular compartment in brain tissue samples.
Taken together, the fate of internalized aptamers is ended up in endocytic vesicles, such as endosomes and lysosomes, and then is redistributed to other subcellular compartments, depending on the physiology of the host cells (Fig. 2 ). But, how internalized aptamers are redistributed to subcellular organelles remains to be investigated.
Fig. 2.
Subcellular localization of internalized aptamers. After internalization, aptamers colocalize with various subcellular compartments, including endosomes, lysosomes, mitochondria, Golgi, and endoplasmic reticulum, depending on their targets.
3.2. Alteration of target protein distribution
The majority of aptamers is antagonistic to their target proteins. Their biologically inhibitory effects have been well-investigated. However, the physico-chemical interactions at protein have not been studied yet. Herein, one compelling study has performed the inhibition of physical movement of targets in cells with anti-nucleolin AS1411 aptamer. The nucleolin is a multifunctional shuttle protein present in the nucleoli, cytoplasm, and plasma membrane [19] and its binding partner is protein arginine methyltransferase 5 (PRMT5) [20]. In free binding statics of AS1411, the nucleolin-PRMT5 complex was distributed in both the nuclear and cytoplasmic compartments. However, in binding statics of AS1411, the nucleolin-PRMT5 complex was decreased in the nucleus and was increased in the cytoplasm of DU145 prostate cancer cells [21]. It suggests that the shuttling function of nucleolin might be affected by the AS1411 aptamer.
4. Fluorogenic aptamers and intracellular imaging
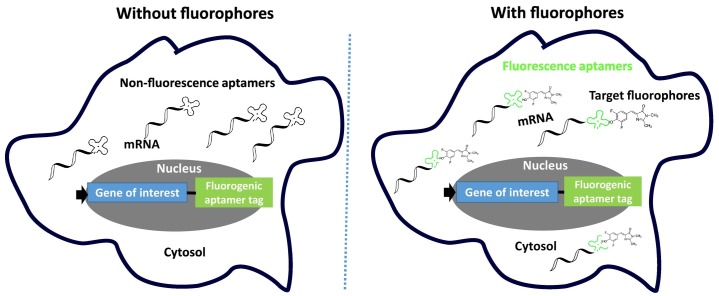
Live cell imaging is valuable for studying the functions and regulation of intracellular RNAs. A recently developed RNA imaging strategy was to use aptamers; Spinach [22], Broccoli [23], and Mango [24] that activated the fluorescence upon binding to their targets of small molecule fluorophores. These fluorogenic aptamers provided a powerful tool to investigate the dynamics of RNA localization and trafficking in cells. As these fluorogenic aptamers have great potentials for a simple protein-free live cell imaging, the details of fluorogenic aptamers and their application are depicted in this section.
4.1. Spinach
The first fluorogenic RNA aptamer ‘Spinach’ which was capable of turning on the fluorescence were isolated upon the binding of small-molecule fluorophores, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), mimicking the green fluorescent protein (GFP) [22]. The Spinach showed the cross fluorogenic activity to the variants of DFHBI, such as DFHBI 1T and DFHBI 2T. In the molecular structure analysis using biochemical probing and secondary structure prediction algorithms, Spinach was predicted to fold into a five-way helical junction [22].
To reveal the mechanism of fluorescence activation by Spinach, the crystallization of Spinach-DFHBI complex was determined. The co-crystal structures of Spinach-DFHBI revealed an unexpected structure composed of a quadruplex core flanked by two coaxially-stacked A-form duplexes [25]. Typically, G-quadruplexes contain guanines in all of their tetrads. However, the quadruplex element of Spinach exhibited unconventional complex connectivity of two G-tetrads and a mixed-base tetrad [25]. The fluorophore was sandwiched by a base triple and was hemmed in by an unpaired guanine residue (G31). In comparison with the crystal structure of DFHBI-free Spinach, the overall conformation of the aptamer was unchanged, but the fluorophore binding site collapsed with a rearranged base tier of the quadruplex and G31 extruded from the helical stack [25]. As the Spinach showed the thermal instability and misfolding that resulted in reduced brightness [26], new variants of Spinach, such as Spinach2 [26], iSpinch [27], and Baby Spinach [28] have been engineered to overcome these shortcomings for further studies.
As the DFHBI fluorophores do not exhibit nonspecific fluorescence and cytotoxicity in cells [22], the feasibility of the Spinach was tested for in vivo imaging. For this, the Spinach was engineered as a genetically-encoded tag to the cellular metabolites and second messenger cyclic di-AMP (cdiA) [29, 30]. For the fluorescence imaging of cellular metabolites, the RNA-based sensors comprising five ligand (adenosine, adenosine di-phosphate (ADP), a-adenosyl-l-methionine (SAM), guanine, or guanosine 5-triphosphate (GTP))-binding RNA aptamers and Spinach via a stem sequences that functioned as a transducer were engineered [30]. The adenosine, ADP, SAM, guanine, and GTP-Spinach sensors showed 20-, 20-, 25-, 32-, and 15-fold increases in fluorescence, upon binding their cognate ligand, respectively [30]. But it showed the rapid fluorescence activation and deactivation kinetics. To monitor metabolic dynamics in live Escherichia coli (E. coli) imaging, SAM-Spinach sensor was used. DFHBI-treated E. coli expressing the SAM-Spinach sensor showed the increased fluorescence ~6 fold over 3 h in the presence of the SAM precursor of methionine [30]. Similarly, ADP-Spinach sensor also detected the dynamic changes in ADP levels which is 20-fold increases in fluorescence upon metabolite binding in E. coli [30].
To visualize intracellular cdiA levels in living Listeria monocytogenes (L. monocytogenes), fluorescent biosensors by combining the ligand-sensing domain of cdiA riboswitch (yuaA riboswitch) with the Spinach2, termed YuaA-Spinach2, were engineered [29]. To test the function of the yuaA-Spinach2 fluorescent biosensor in live cells, plasmid encoding the biosensor in a tRNA scaffold was transformed into L. monocytogenes. In results, the cellular fluorescence was observed by the fluorescence microscopy and flow cytometry after incubation with DFHBI [29]. The biosensor tagged with the Spinach and Spinach 2 showed the better imaging in living E. coli and L. monocytogenes cells [29].
4.2. Broccoli
Another green fluorogenic RNA aptamer ‘Broccoli’ which was binding to the fluorophore DFHBI was also isolated by the same group that developed the Spinach [23]. The Broccoli was isolated using the combined methods which were the systematic evolution of ligands by exponential enrichment (SELEX) and fluorescence-activated cell sorting (FACS) approaches. The SELEX was performed in the first round when the RNA pool exhibited fluorescence upon incubation with the fluorophore. After that, the aptamer library was cloned into a bacterial expression plasmid. The library plasmid was transformed into E. coli and in vivo fluorescence was sorted out by FACS in the presence of DFHBI. The characteristics of the Broccoli showed robust folding in low magnesium concentrations, as well as increased thermostability and fluorescence than Spinach2. The Broccoli was engineered to tBroccoli (49 nt) and the dimeric tBroccoli (tdBroccoli) for further in vivo applications. Indeed, the tdBroccoli showed the twice as fluorescence as Broccoli in living E. coli [23]. For the genetic expression of structured RNAs, the tRNA-fused scaffold system is often used to promote the appropriate folding of aptamers in cells [31], because the appropriate structural folding of aptamers is pivotal for the function of aptamers. For the stable folding of aptamers, the tBroccoli or tDBroccoli was fused to the 3′ terminus of 5S expressed from the pAV5S tRNA scaffold plasmid (5S-tBroccoli or 5S-tDBroccoli) for imaging in mammalian HEK293T cells. The 5S-tdBroccoli showed 70% brighter than 5S-tBroccoli in mean fluorescence intensity in transfected HEK293T cells. Moreover, the Broccoli and dBroccoli fused to the 3′ terminus of 5S without tRNA scaffold plasmid showed the higher cellular brightness than 5S fused to tRNA scaffold-aptamer constructs in HEK293T cells. It supports the idea that the tRNA scaffold is selectively necessary depending on the folding efficiency of aptamers for genetically tagged imaging.
4.3. Mango
The RNA aptamer ‘Mango’ binding to derivatives of fluorophore thiazole orange 1 (TO1) was developed in the series of fluorogenic aptamers [24]. In the molecular structure analysis using the sequence analyses and biochemical probing, the Mango was predicted to fold into two-tiered all-parallel G-quadruplexes connected to an A-form duplex [24]. To reveal the mechanism of fluorescence activation by the Mango, the crystallization of Mango-TO1 complex was determined. The co-crystal structure of Mango-TO1 revealed a three-tiered G-quadruplex that included three antiparallel quinine residues connected to an A-form duplex through a GAAA tetraloop-like junction [32]. The fluorophore bound on one of the flat faces of the G-quadruplex, stabilized the Mango-TO binding for fluorescence. Therefore, the Mango enhanced the fluorescence of the fluorophore up to 1100-fold relative to unbound fluorophore. As the Mango showed the brightest fluorescence among other Spinach and Broccoli fluorogenic aptamers, the feasibility of in vivo imaging was determined in Caenorhabditis elegans (C. elegans). To visualize the cellular RNAs with the Mango, TO1 was directly injected into C. elegans syncytial gonads, either with or without RNA Mango. The injection of dye alone showed the negligible fluorescence, but the injection of the Mango resulted in a strong fluorescence, specifically localized to the syncytial nuclei of the gonad [24]. The unbleached bright fluorescence was observed over 2 h after injection within time frame in vivo.
Given that the Mango showed the rigid fluorescence binding to target molecules and only fluorogenic aptamers working in vivo model, the optimization of the Mango are expected to appear for in vivo mammalian imaging.
4.4. Corn
The RNA aptamer ‘Corn’ which activated the yellow fluorescence on the binding of 3,5-difluoro-4-hydroxynenzylidene imidazolinone-2-oxime (DFHO) was developed by the same group that isolated the Spinach and the Broccoli [33]. The Corn showed improved photostability, compared to Spinach and Broccoli, which enables quantifying RNA transcription in living cells. As for a genetically encoded fluorescent RNA reporter, the Corn in a tRNALys scaffold was expressed in three main subclass of Pol III promoters, 5S RNA, tRNA, and U6 promoters. The transfection of these reporter plasmids into HEK293T cells led to yellow fluorescence after addition of DFHO. But, in the treatment of actinomycin D (an inhibitor of RNA polymerase), the reduction of yellow fluorescence was observed [33]. Although the Corn showed the enhanced photostability, its dimerization makes unsuitable for imaging tags in cells due to the structural changes [33].
4.5. New Mango
The new high-binding RNA Mango fluorogenic aptamers have been isolated based on the microfluidics-based selection methods [34]. The newly isolated Mangos showed as bright as or brighter than enhanced green fluorescence protein (EGFP) when bound to their target small molecule TO1. To test the cellular imaging of new Mango-tagged RNAs, the subcellular localization of two small non-coding RNA (5S and U6) was investigated in mammalian cells. For the imaging of 5S, the human 5S ribosomal RNA was tagged with new Mango by incorporating an F30 folding scaffold. The in vitro-transcribed 5S-F30-Mango RNA was transfected into HEK293T cells. It showed up to 10 bright RNA foci per cells. The majority of 5S-F30-Mango foci were cytoplasmic and a small faction in nuclear. In the subcellular localization studies, the 5S-F30-Mango foci overlapped with mitochondria, not with other subcellular compartments such as P-bodies, endosomes or stress granules. To confirm the observation were specific, the U6 snRNA (small nuclear RNA) which is associated with ribonuclear protein (snRNP) were tagged with new Mango in F30 folding scaffold. The transfected in vitro-transcribed U6S-F30-Mango RNA showed 9 fold increased foci in nuclear. The U6-F30-Mango is was mainly overlapped with small nuclear protein (snRNP) Lsm3, not mitochondria or ribosomes [34]. These results highlight the new Mango tag can be used to track the preferential cellular locating of small cellular RNAs.
For the function as genetically encoded tags expressed in cells, the plasmid was constructed to express 5S rRNA under RNA pol III promoter in pSLQ plasmid. The pSLQ-5S-F30-Mango construct exhibited an increased fluorescent signal in nucleolar compartments as well as cytoplasmic foci, demonstrating that the capability of new Mango tags to function as efficient genetically encoded reports of RNAs. But the subcellular localization was limited on ribosomal protein L7, not on mitochondria. These results showed the inconsistent colocalization, compared with the transfection of in vitro-transcribed 5S-F30-Mango RNAs.
Currently, live RNA imaging in eukaryotic cells is a big challenge with these fluorescence aptamers due to the main issues; the low intensity, rapid photo bleaching and aptamer's structural changes. Hence, very few literatures have shown the successful imaging in mammalian cells. In the comparison of fluorophore ligand binding and complex stabilization studies, the Mango has a large thermal stability and better discrimination from a broad range of small molecules [35] and the Corn shows improved photostability. Therefore, there are no doubt that the Mango and the Corn could be good tools to elucidate the working mechanism of cancers biology fields.
4.6. Imaging of structured viral RNAs
In viral diseases, to investigate viral RNA dynamics, the full cDNA of Sindbis virus (SINV) was constructed with two copies of tRNA-scaffolded Spinach2 [36]. Live cell imaging revealed that the Spinach2-tagged SINV viral RNAs mainly localized to the tip of filopodia of infected cells where they contact adjacent non-infected cells. Later, viral RNA was observed at the contact site of uninfected cells and then spread to the cytosol. This study clearly demonstrates that fluorogenic aptamer tag might be a good tool to investigate the dynamic of viral RNAs in virology fields.
4.7. Aptamer-based Förster resonance energy transfer (apta-FRET)
For the molecular interactions in living cells, two fluorescent RNA aptamers, Spinach and Mango, were engineered using RNA origami scaffolds for detection of conformational changes, called aptamers-based Förster resonance energy transfer (apta-FRET) [37]. To obtain FRET, the Spinach and Mango were placed in close proximity on the scaffolds and genetically encoded in the plasmid. The apta-FRET constructs expressed in E. coli showed the tolerable FRET output [37], suggesting that the apta-FRET system might be used as real-time imaging in living cells and biosensors.
4.8. Summary
Fluorogenic RNA aptamers, such as Spinach, Broccoli, Mango, Corn, and new Mango are very powerful tools that can be genetically encoded tags with a gene of interest for live-cell imaging. But, still remaining issues are how well tolerated when they are engineered. To get the answer, one study was performed how well these fluorogenic RNA aptamers are tolerated when engineered for structured cellular RNAs [38]. Nine distinct variants of Spinach and Broccoli were inserted at the same position in 16S ribosomal RNA (rRNA) for comparative studies. Among the nine variants, Baby Spinach and Broccoli yielded superior fluorescence in live-cell imaging of E. coli. However, most variants of Spinach and Broccoli impaired cell growth depending on the sequences and length of the aptamers, showing the toxicity. Hence, the optimization is indispensable. As all of strategies described here enables the design of sensors to image essentially any molecules in living cells, the fluorogenic RNA aptamer tags have diverse applications for imaging of intracellular RNA dynamics (Fig. 3 ). Also, theoretically, RNA aptamers can be readily generated against any biomolecules, new fluorogenic aptamers are expected to be developed.
Fig. 3.
Intracellular imaging with fluorescent aptamers. The fluorogenic RNA aptamers, turning on the fluorescence on binding of their target fluorophores, are genetically engineered into the gene of interest as imaging tags. After translation, fluorogenic RNA aptamers can visualize the dynamics of intracellular genes with fluorophores in live cells.
5. RNA aptamers for regulating gene expression in E. coli
Sequence-specific DNA-binding proteins called transcription factors (TFs) regulate transcription activation or repression, playing a critical role in the control of gene expression in living organisms [39, 40]. For the proof of concept studies, RNA aptamers have been developed to target the TFs to regulate the gene expression in both prokaryotes and eukaryotes. As the regulation of gene expression by aptamers is a very promising therapeutic strategy, the representative examples of gene regulation by aptamers are depicted in this section.
5.1. Transcription-activating aptamers in E. coli
Artificial RNA aptamers have promising roles as non-coding regulatory RNAs used for gene regulation and molecular engineering. Over the last decade, the functional role of regulatory RNAs in gene expression has been well developed in prokaryotes, specifically transcriptional activation of gene expression by aptamers in E. coli. The RNA aptamers against the bacterial transcriptional regulator Tet repressor (TetR) activated transcription by modulating the DNA-binding activity of the TetR [41]. The activity of intracellularly expressed TetR aptamers directly correlated with its expression level and stability. The same group, again, isolated inhibitory RNA aptamers against the repressor protein lambda phage cI as a transcriptional activator in E. coli [42]. Upon intracellular aptamer expression, expression of the reporter gene was increased 35-fold. These studies illustrate valuable strategies for the fine regulation of genetic circuits by using synthetic RNA aptamers.
Conventionally, artificial aptamers are isolated against their targets with a synthetic aptamer library pool. To systematically search for natural RNA aptamers that regulate transcription through direct interaction with RNA polymerase (RNAP) in bacteria, genomic SELEX; a genomic library of E. coli as a source of RNA sequences and the E. coli RNAP holoenzyme as bait, were performed [43]. Following the next-generation sequencing and functional analysis, 15,000 of natural RNA aptamers to RNAP (RAPs) were identified in the E. coli genome. The majority of identified RAPs (64.3%) was characterized inhibitory RAPs (iRAPs) that encoded in the strand opposite to the annotated genes acting like transcriptional ‘anti-interference’. The antisense iRAPs curbed transcriptional interference by cis-acting as pro-termination signals, potentiating the expression of sense-encoded genes [43]. This study shows that natural RNA aptamers have potential as a self-regulatory system.
5.2. Conditional control of gene expression
Robust synthetic tools are useful for the control of cellular gene regulation. As a proof-of-concept study for ligand-mediated ribozyme cleavage, a tetracycline (Tet) aptamer was grafted to the hammerhead ribozyme and inserted into the 3′ untranslated region (UTR) of reporter genes [44]. The working mechanism was that a ligand binding to the aptamers destroyed a loop–loop interaction within the ribozyme thereby inhibiting ribozyme cleavage and allowing gene expression. For the function assay, the Tet-aptamer-ribozyme vector was transfected into HeLa cells. The cells expressing the Tet-dependent ribozymes showed increased expression of the reporter genes up to 8.7 folds in the presence of Tet, but not in absence [44].
As a proof-of principle concept, conditional control of gene expression by Tet-aptamer-ribozymes has been successfully established in mammalian cells.
6. Transcription factor (TF)-targeting aptamers
Transcription factors (TFs) have pivotal roles in regulating gene expression during cell growth, development, and cellular homeostasis. Therefore, the dysregulation of TFs is linked to many types of diseases, which makes them attractive targets for therapeutic development. Several natural prokaryotic and eukaryotic RNAs that modulate the activity of TFs such as 5S RNA, 6S RNA, 7SK, hepatitis delta virus RNA, neuron restrictive silence element RNA, growth arrest-specific 5, have been already reported [45]. As nucleic acid aptamers can be encoded in transgenes for endogenous expression, transcription-regulating aptamers have unique advantages for therapeutics. Hence, the intriguing concept of synthetic aptamers that target DNA-binding TFs is depicted, with an emphasis on emerging cancer therapeutics.
6.1. Anti-forkhead box M1(FOXM1) aptamers
The transcription factor FOXM1 is a member of the fork head/winged-helix (Fox) family that activates a network of proliferation-associated genes and is frequently overexpressed in cancers [46]. FOXM1 contains a highly conserved DNA binding domain (DBD); therefore, targeting the DBD of FOXM1 could suppress its transcriptional activities and presents a potent therapeutic strategy in cancer [47]. The DNA aptamer targeting the FOXM1 DBD (FOXM1 Apt) was isolated and phosphorthioate-modified FOXM1 Apt (M-FOXM1 Apt) was constructed to increase its serum stability [48]. Both FOXM1 Apt and M-FOXM1 Apt showed specific binding to FOXM1 proteins and M-FOXM1 Apt improved the binding affinity from 172.1 nM to 63.86 nM. M-FOXM1 Apt localized to the nuclei of cells after transfection. To test the inhibition of transcriptional activities of M-FOXM1 in vitro, gel shift assay was performed. It abolished the binding of FOXM1 on its consensus binding sites at a dose-dependent manner. In biological functional assays, M-FOXM1 Apt significantly inhibited cell proliferation at the 48 h time point after treatment. The treatment of M-FOXM1 Apt dramatically decreased the expression of FOXM1-target genes, such as Cdc25B and Cycline B1, but not FOXM1 in breast cancer cell. It suggests that the inhibitory effects of M-FOXM1 result from disturbing FOXM1 transcriptional activation but not from directly affecting FOXM1 expression [48].
6.2. Anti-heat shock factor (HSP) aptamers
The Heat shock factor (HSF1) is a TF that regulates chaperone protein expression for cell survival under stress conditions, particularly in malignant cancers [49]. Anti-HSF1 aptamers isolated against Drosophila HSF1 proteins (iaRNAHSF1) was delivered into HeLa cervical carcinoma cells in the form of a synthetic gene containing a hammerhead ribozyme [50, 51]. The iaRNAHSF1 expression induced morphological changes and apoptosis in cervical cancer cells but not in normal cells, showing its cancer specific effects. Levels of various molecular chaperone proteins such as heat shock protein 70, calnexin, transglutaminase 2, and glucose-regulated protein 78, were also significantly decreased in aptamer-expressing cells, suggesting that the inhibition of molecular chaperons by iaRNAHSF1 is due to a general decrease in HSF1 activity on its gene targets.
6.3. Anti-nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) aptamers
The transcription factor NF-kB is an important activator of gene involved in immune functions such as inflammation, the synthesis of cytokines and interferons, apoptosis, and cancers [52, 53]. Hence, the inhibition of NF-kB dependent gene activation is one of good strategies for enhancing the activities of anticancer drugs. For the soluble molecular decoys of NF-kB, RNA aptamers was identified against the p50 homodimer form of NF-kB (p502). In vitro assays, the selected anti-NF-kB aptamers competitively inhibited DNA binding by NF-kB [54]. The following studies have been performed to optimize the RNA aptamer for binding to NF-kB [55]. The optimized anti-NF-kB aptamers demonstrated the specific decoy activity, inhibiting transcriptional activation by its NF-kB target protein. In the expression of bivalent of anti-NF-kB p50 aptamers, the decoy activity was enhanced [55]. This study has a strong implication that the expression of therapeutic decoy RNAs have possibility to modulate gene expression. The anti-NF-kB p65 was also isolated by the same group in parallel [56]. The selected anti-NF-kB p65 aptamers had the similar decoys effect in competition assay. Hence, the potential therapeutic strategy is to combine the p50 and p65 aptamers to form a heterodimeric aptamer that is competent for simultaneous binding of both subunits of NF-kB heterodimers.
6.4. Anti-runt-related transcription factor 1 (RUNX1) aptamer
The transcription factor RUNX1 is an important regulator of hematopoiesis and important partner in leukemia [57, 58]. For the leukemia therapeutics, RNA aptamers were isolated to the recombinant RUNX1 Runt-Homology-Domain (RHD) and its interaction partner, the core-binding factor β protein (CBFβ) [59]. In competition experiments, the identified anti-RHD-CBFβ aptamers disrupted the formation of DNA-RHD-CBFβ complexes, suggesting that the RNA aptamers could act as molecular DNA mimics, adopting three-dimensional structures with similar physico-chemical properties to cognate RHD DNA.
7. Therapeutic intracellular targeting aptamers (intramers) and drug screening
The aptamers have been developed against the cell surface receptors for the targeted therapeutics and delivery [60]. Their successful stories are well constructed in the field of therapeutic development in viral disease and cancers. But, rather than the cell surface receptor aptamers, intracellular targeting aptamers (intramers) are depicted for the new fields of therapeutic development in this section.
7.1. Intramers in viral diseases
Therapeutic intramers have been actively developed against viral proteins such as human immunodeficiency virus (HIV) Tat [61] and reverse transcriptase [62], hepatitis C virus NS3 protease/helicase [63, 64], NS5B RNA-dependent RNA polymerase [65], and severe acute respiratory syndrome NTPase/helicase [66]. But mutation of the viruses changes the structure of these target proteins, nullifying the effects of anti-viral drugs. Accordingly, anti-viral drug development is moving toward more highly conserved targets such as viral UTR regions.
HIV-1 is a single-stranded RNA lentivirus of the family Retroviridae. Its genome is 9.2 kb with multiple open reading frames, flanked by a UTR at both the 5′ and 3′ ends [67]. For the universal anti-viral effects, RNA aptamers have been isolated targeting the first 308 nt of the 5′-UTR, which is highly conserved in HIV-1 [68]. After 14 rounds of selection, the 64-nt length of anti-HIV-1 UTR aptamer was isolated. In the sequence analysis, the hexanucleotides (5′-GGCAAG-3′) within anti-HIV UTR aptamers were found to be complementary to the apical loop of the poly (A) domain in the repeated region (R) located in both the 5′- and 3′-UTRs of HIV, indicating that the hexanucleotide sequence is a putative binding site. Use of in silico aptamers-target analysis, anti-HIV UTR aptamer was truncated to the 16 nt-long stem-loop RNA aptamers including the hexnucleotides (RNApt16). But, for the function assays, the RNApt16 and unmodified full-length of anti-HIV UTR aptamers did not show any inhibitory effects with transfection. But, chemically synthesized RNApt16 showed inhibitory effects of 85 ± 5%, suggesting that improving the half-life is pivotal. By following studies, the full length anti-HIV UTR aptamers were cloned into a human U6 snRNA cassette, pU614, for stable hairpin-loop domains. HEK293T cells were transfected with the in vitro transcribed anti-HIV UTR aptamers from the U6 plasmid, which inhibited HIV particle production upto 80 ± 7%.
7.2. Intramers in aging and age-related diseases
7.2.1. Alzheimer's disease (AD)
Proteasomes have a critical role in protein homeostasis regulation via ubiquitin (Ub)-dependent proteolysis when proteins are intrinsically disordered [69]. The Ub-proteasome system has its own quality control mechanisms, one of which is the proteasome-associated deubiquitinating enzyme ubiquitin carboxyl-terminal hydrolase 14 (USP14). Deletion or small molecule inhibition of USP14 results in accelerated proteasomal degradation of various target substrates [70], suggesting that USP14 is a therapeutic target for treating diseases caused by the accumulation of damaged and misfolded proteins, such as neurodegenerative diseases. In one study, anti-USP14 RNA aptamers were isolated based on the protein SELEX and found to facilitate the degradation of Alzheimer's disease-implicated Tau proteins via enhanced proteolytic activities of proteomes and inhibition of deubiquitinating activity. This results suggests that USP14 aptamers as a promising therapeutic option for Alzheimer's disease [71].
7.2.2. Diabetic nephropathy
An extracellular matrix protein of periostin is a secreted proteins that functions as both a cell attachment protein and an autocrine or paracrine factor that signals through the cell adhesion molecules αvβ3 and αvβ5 integrin [72]. Pathophysiologically, it involves wound repair [73] and tumorigenesis [74]. Originally, anti-peripstin DNA aptamers developed to inhibit breast cancer growth and metastasis [75]. But, recently accumulated evidence suggests that the high expression of periostin is involved in renal fibrosis through the hyper activation of transforming growth factor-β1 (TGF-β1) pathway [76, 77]. Indeed, the accumulation of periostin is associated with progressive renal fibrosis in diabetic nephropathy by the activation of TGF-β1, which is regarded as one of the main mediators of the deleterious effects of diabetic nephropathy [78, 79]. To determine the feasibility of anti-periostin DNA aptamers for the therapeutics for renal fibrosis in diabetic nephropathy, the biological function of anti-periostin DNA aptamers was determined in diabetes mellitus animal model. It showed that anti-periostin DNA aptamers attenuated increased blood urea nitrogen levels and expression of fibronectin and tubule interstitial fibrosis under diabetic conditions in vivo [80], suggesting that anti-periostin DNA aptamers might be a potential therapeutic to treat fibrosis in diabetics.
The periostin also promotes hepatic fibrosis by modulating hepatic stellate cell activation via αv integrin interaction in mice [81]. Hence, anti-periostin DNA aptamers have implication for the applicable therapeutics to treat liver fibrosis.
7.2.3. Cancers
Identifying cancer specific aptamers is pivotal to increase the therapeutic index for cancers. The recently developed cancer specific aptamers targeting cancer plasma membrane have been well functionalized to delivery therapeutic small RNAs, small molecules, antibodies, and peptides, even novel cancer specific biomarker discoveries [82]. The RNA aptamers targeting the cancer stem cell (CSC) surface marker CD133 [83] and other markers such as CD117, EpCAM, prostate CSC, HER2, and MRP1 have been well characterized for the cancer specific therapeutic effects [84]. Along with these aptamers, cancer specific point mutations and isotypes are very attractive targets for the specific anti-cancer effects. Herein, the representative cancer specific intramers are depicted.
Protein with a single amino acid substitution is the cause of a plethora of human diseases [85]. Also, point mutations in multiple tumor suppressor proteins cause cancers [86] Actually, >50% of human cancers is related to mutations in p53, a tumor suppressor, and the single amino acid substitution p53R175H is one of the mutations at the p53R175 which is a dominant mutation of p53 [87]. The p53R175H mutation abolished the function of the p53 wide type (WT) and confers a gain of anti-apoptotic effects in lung cancer cells [88]. To select p53R175H specific RNA aptamers, contrast screen SELEX was used with WT p53 proteins. RNA aptamers binding to WT p53 was discarded and the RNAs binding to the p53R175H were selectively recovered and amplified. After five round of SELEX, RNA aptamers that bind specifically to p53R175H have been successfully isolated [89]. The specific binding to p53R175H, but not the WT p53 was confirmed by gel shift assays. Further in vitro function assay, intracellular delivery of anti-p53R175H aptamers with nanoparticles did not affect cell growth harboring WT p53 and another type of p53 mutant, p53R273H. But, anti-p53R175H aptamers clearly showed the inhibition of cell growth harboring p53R175H. For the further lung tumor cell xenografted animal model, p53R175H mutant harboring tumor cells were subcutaneously injected to the nude mice. The p53R175H-RNA aptamers coated with nanoparticles were injected two times at day 5 and day 8 by subcutaneously to the tumor. It showed significantly slower lung tumor growth by inducing the apoptosis [89].
β-arrestins, the ubiquitous cellular scaffolding proteins, have been considered attractive therapeutic targets because of their role as signaling mediators in critical tumorigenesis pathways [90]. Two isoforms of β-arrestins, β-arrestin 1 and 2, sharing high sequence homology and similar structures have distinct roles in signaling [91]. The genetic ablation of β-arrestin 2 protects animal from the progression of myeloid leukemia [92]. For the therapeutic purpose, β-arrestin 2 specific RNA aptamers were isolated with counter SELEX against β-arrestin 1 [93]. The anti-β-arrestin 2 aptamer showed the high binding affinity to β-arrestin 2, but 35–500 fold lower binding affinity to β-arrestin 1. For the intracellular delivery strategy, the DNA ‘targeting aptamer’ which is nucleolin aptamers were linked with ‘therapeutic aptamer’ of anti-β-arrestin 2 aptamers by complementary base pair annealing. This nucleolin-β-arrestin 2 aptamer chimeras allowed delivering intramers in chronic myelogenous leukemia cells without any viral vector or transfection agents. The nucleolin-β-arrestin 2 aptamer chimeras showed fully functional inhibition of β-arrestin 2 signaling cascades, showing down-regulation of β-catenin and Gli [93]. Consequently, the nucleolin-β-arrestin 2 aptamer chimeras inhibited the leukemia cell growth by colony forming assays [93]. The anti-tumor effects were not determined in vivo.
The translational dysregulation for the increased cellular proliferation and pro-oncogenic mRNAs is a hallmark of many cancers [94, 95]. Such mRNAs hold the high structured 5′UTRs requiring high levels of eukaryotic initiation factor 4A (eIF4A) helicase activity for efficient translation [96]. The recent observation of increased eukaryotic initiation factor 4A (eIF4A) activity in the breast cancer malignancy [97] also prove that mRNAs are most dependent on eIF4A1 activity in cellular transformation, which is making eIF4A an attractive target for therapeutic intervention. Indeed, RNA aptamers to eIF4A have been characterized using protein-based SELEX methodology [98]. In vitro functional assays, anti-eIF4A showed the inhibition of cap-dependent translation by blocking ATP hydrolysis [98]. In the working mechanism of anti-eIF4A aptamer, it was proposed that anti-eIF4A aptamers inhibited the conformation changes necessary for ATP hydrolysis. Based on the fact that the malignant cells become ‘addicted’ to the altered translational landscape [99], the selected anti-eIF4A aptamer has a great potential as therapeutics against all type of malignant cancers.
The still remaining challenges of intramers for therapeutic options are delivery. But, the above two studies clearly showed the successfully delivery of intramers into the cells using nanoparticles or aptamers. Hence, aptamers targeting intracellular antigens are promising therapeutic fields to be expanded in cancers.
7.3. Intramers for drug screening
Traditional approach to study the function of a given protein in the living cells is based on the small molecule drug that exhibits high specificity, affinity, and inhibitory activity. The majority aptamers show the inhibitory of protein function. Therefore, one intriguing idea was brought up that aptamer/protein complex with verified intracellular functionality was used to develop assays to screen small-molecule libraries [100]. The main concept was to displace the aptamer from its target protein, releasing the fluorescence signals if a small molecule competes with the aptamer/protein interaction. For the high-throughput screening, reporter ribozyme was developed to screen a 96 member sample library of antibiotics for molecules that could disrupt the interaction between HIV Rev and its cognate RNA. If small molecules compete with the binding site of the aptamer/protein complexes, the ribozyme become active and cleaves a substrate labeled with a fluorescence tag and a quencher. In the cleaved state, the two products rapidly dissociate from the ribozyme, resulting in a fluorescence signal. In results, the screen identified one compounds, coumermycin A1, that inhibited the HIV-1 virus replication in cell culture experiments [100]. This proof of concept study well establishes the novel small drug inhibitor screen assays by using interference with aptamer/protein interactions.
8. Conformation-selective aptamers
8.1. Allosterically modulating aptamers
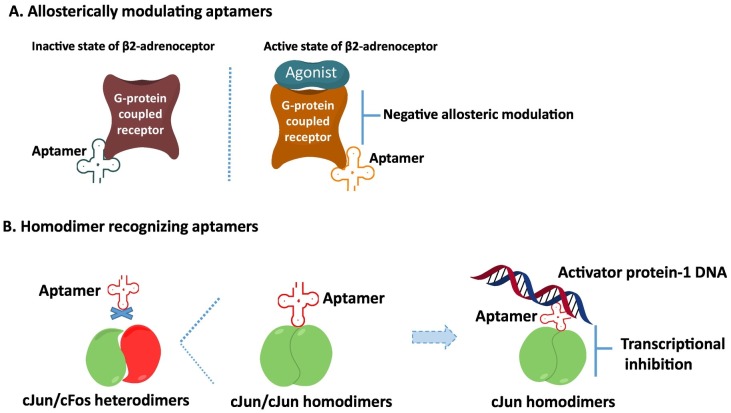
Current molecular biology techniques have the limited ability to distinguish the conformational changes of proteins. Aptamer's superior target recognition function allows us to use as a tool to detect the conformation changes of target proteins. Indeed, a very compelling study has been investigated how sophistically aptamers can detect the conformational changes and how it affect the downstream signaling in G-protein-coupled receptors (GPCRs). GPCRs are the seven-α-helical transmembrane-spanning receptors and over 800 members are identified in the human genome [101]. GPCRs are very popular targets in pharmaceutic industries and one-third of pharmaceutical agents targeting GPCRs are currently available on the market for the treatment of various human diseases [101]. The GPCRs undergo conformational changes that activate intracellular signaling cascades in response to agonist binding. It exhibits conformational heterogeneity in both ligand-occupied and ligand-free states [102, 103]. To take the full advantage of aptamer's high target recognition to detect the conformational change of GPCRs by binding of agonist, a highly diverse RNA library combined with the next-generation sequencing and bioinformatics analyses were used to isolated RNA aptamers that bound a prototypical GPCR, the β2-adrenoceptor (β2AR) [104]. For the SELEX, β2AR proteins were produced by baculovirus-mediated expression in Spodoptera frugiperda 9 (Sf9) insect cells. To keep the active conformation state of β2AR for selection of RNA aptamers, an agonist of BI167107 was used. In results, several aptamers were grouped into three categories: 1) conformational selective aptamers (A1, A2, and A13) for the active form of the β2AR on the BI167107-bound; 2) binding specific aptamers (A15 and A16) for inactive form of the β2AR; 3) clear selective aptamers (A11) binding to both unoccupied and BI167107-bound forms of the β2AR [104]. For further characterization, the modulation of transitions between active and inactive conformation as well as the stabilization of the ligand-specific conformation was explored using agonists, antagonists, and inverse agonists. Comparing with the control, aptamers A1 and A13 robustly bound to agonist-occupied β2AR which was correlated with agonist efficacy, and this binding was markedly reduced in the presence of antagonists. But, interestingly, the A2 aptamer did not correlate with ligand efficacy. To investing the working mechanism, the receptor pulldown assay with BI167107-occupied β2AR was performed. Surprisingly, the aptamer A2 displayed a unique selectivity toward a β2AR conformation stabilized by inverse agonists, suggesting that the aptamer A2 stabilizes a unique active conformation of the β2AR that is distinct from that stabilized by aptamers A1 and A13. To investigate the ability of aptamers to stabilize active β2AR conformations, the fluorescence spectroscopic studies (that is, the decreased fluorescence intensity upon the conformational changes) were performed with a β2AR labeled with a bimane probe. The results showed the enhanced effects of A1, A2, and A13 (that is, a further decrease in fluorescence intensity) when they were combined with full agonist, signifying further stabilization of active conformations by aptamers. To investigate the binding epitopes and structure basis of the interactions, negative stain electron microscopy (EM) and single-particle reconstruction analysis was utilized. The visualization of aptamer-β2AR complex showed that A16 appeared to interact with the extracellular region of the receptor. On the contrary, the A1, A2, and A13 bound at the intracellular region of the β2AR, suggesting that binding of the aptamers to the active state of β2AR inhibited the receptor in the presence of an agonist by engaging an allosteric mechanism [104] (Fig. 4A). It indicates the aptamers might be used allosteric inhibiters. There was reported no relationship between binding affinity and the functional effects on β2AR aptamers. This study supports the idea that the binding site of the aptamer on targets, and not binding affinity, is important for functional effects.
Fig. 4.
Conformational change recognizing aptamers. (A) Allosterically modulating aptamers. RNA aptamers that specifically recognize the active state of β2-adrenoceptor (β2AR) act as allosteric modulators of the G-protein-coupled receptors (GPCR), binding at distinct sites to inhibit its signaling activity. (B) Homodimer-recognizing aptamers. DNA aptamers that are >100-fold specific for binding cJun/cJun as compared to cJun/cFos block binding of the transcription factor activator protein 1 (AP-1) DNA element and inhibit cJun homodimer transcription in cells.
8.2. Conformation-specific aptamers
Heat shock protein 70 (Hsp70) is a ubiquitous molecular chaperone that plays a critical role in proteostasis. Driven by ATPase activity, Hsp70 holds two conformations, Hsp70-ATP and Hsp70-ADP [105]. Conformation-specific RNA aptamers against Hsp70-ATP, but not Hsp70-ADP, have been isolated [106]. The anti-Hsp70-ATP aptamers inhibited the ATPase activity of Hsp70 during ATP hydrolysis. As Hsp70 is increasingly being recognized as a drug target in a number of age related diseases such as neurodegenerative, protein misfolding diseases and cancers, anti-Hsp70-ATP specific aptamers have great potential in therapeutic applications.
The activating protein-1 (AP-1) family of transcriptional factors consists of homodimers and heterodimers of Jun or Fos that regulate a variety of transcriptional processes in response to stimuli and recognize the AP-1 DNA binding site [107]. The different homodimer and heterodimer compositions of Jun and Fos show promoter-specific differences in transcription activation [108]. These AP-1 dimers regulate in variable cellular pathways including cell proliferation, apoptosis and tumorigenesis [109]. However, current molecular biology techniques are unable to distinguish the difference between the roles of cJun/cJun homodimers versus cJun/cFos heterodimers. To build the foundation to investigate the different AP-1 dimer compositions, DNA aptamers have been isolated using the cJun homodimer protein based SELEX [110]. The binding specificity of DNA aptamers to homodimers of cJun/cJun, but not to cJun/cFos heterodimers was confirmed with the gel shift assays. In competition assay, the anti-cJun aptamer blocked the homodimerization of cJun from binding AP-1 DNA. The cJun homodimers are not only capable of binding cis-elements on DNA to activate transcription but can also function as transcriptional co-activators by binding directly to other transcription factors NFATc2 for synergistic activation of interleukin 2 (IL-2) [111]. Therefore to determine the biological function of anti-cJun homodimer DNA aptamers, the transcriptional inhibition of IL-2 was assayed. The anti-cJun dimer aptamer repressed the expression of IL-2 in luciferase assay [110] (Fig. 4B).
8.3. G-quadruplexes in aptamer structures
The specificity of aptamers reflects the arrangement of loops and stems and the hydrogen bonds that stabilize their three-dimensional structures. The structural stability of aptamers relies on a core formed by a stacked G-quadruplex. G-quadruplexes are guanine-rich sequences with extremely stable secondary structures. G-quadruplexes are formed when guanines are organized into planar quartets in which each base is connected to two other bases via Hoogsteen base pairing. Hydrogen bonds between each pair of guanines involve four donor/acceptor atoms, so G-quadruplexes have eight hydrogen bonds in total [112]. However, in some cases, very stable G-quadruplex aptamers have shown cross-binding to several different proteins. For example, the 16-mer G-quadruplex DNA aptamer binds HIV-integrase, interleukin-6, and HIV-gp120 [113, 114]. The G-quadruplex RNA aptamers showed cross-binding to VEGF165, PDGF-AA, and PDGF-BB [115]. The structural stability of aptamers, mainly via G-quadruplex formation, is very important, but it may reduce the specificity of aptamers in some cases. Thus, the G-quadruplex required for aptamer stability has been considered a double-edged sword.
9. Immune responses to chemically-modified aptamers
Recently, concerns regarding potential immune stimulation by synthetic DNA aptamers in human blood have been raised [116]. To avoid the immune stimulation, numerous types of non-natural chemical modifications have been developed for the functional diversity of DNAs, xenonucleic acid (XNA, artificial genetic polymers) [117], and RNAs [118]. Currently, widely used chemical modification in RNA aptamers are 2′O-methyl (2′OMe) and 2′Fluoro (2′F). To investigate the immune responses of chemically-modified RNA aptamers in vitro level, 2′OMe and 2′F nucleotides were incorporated into RNA aptamers. The 2′F modification abrogated the stimulation of toll-like receptors 3 and 7, but enhanced the activity of retinoic acid-inducible gene 1 [119]. The 2′F modification with 5′ triphosphate (5′ppp) increased cell death and interferon-β expression in human cancer cells compared with 2′ hydroxyl with 5′ppp, whereas chemically synthesized 2′F RNA aptamers without 5′ppp did not. Interestingly, replacement of 2′F with 2′OMe in 5′ppp RNA aptamers completely abolished the expression of IFN-β and inflammatory cytokines, suggesting that the main immune response of RNA aptamers is due to 5′ppp. [119].
The most commonly used chemical modification during SELEX is 2′F by in vitro transcription. Indeed, generation of 2′F-5′ppp RNA aptamers during SELEX is unavoidable. Hence to avoid the immune responses: first, after the SELEX, chemically synthesized aptamers should be used for in vivo assays and clinical trials. Second, 2′OMe modification might be considered during the SELEX; but, unfortunately, there are no currently available polymerases to incorporate 2′OMe during the SELEX.
To increase the pharmacokinetics, PEGylation of the aptamers is an often used strategy. In the clinical trial with Pegnivacogin (PEGylated anti-coagulation factor IXa RNA aptamers), the adverse immune responses were found [120]. But, it was later discovered that the reported severe allergic reactions was due to preexisting antibodies to PEG [121].
10. Conclusions and future perspectives
Over the past decades, the great efforts are made to investigate the endocytic mechanism of aptamers and their subcellular distribution in the molecular and cellular level. The main endocytic mechanisms of aptamers are categorized into the clathrin-dependent or –independent, depending on the type of targets or cells. The variations of intracellular distributions are also observed depending on the type of cells. Upon the binding of aptamers to their intracellular targets, antagonistic aptamers induce the alternation of intracellular distribution of their targets and the inhibition of biological functions, suggesting the potential of intracellular therapeutic applications. Indeed, there have been exciting improvements in intracellular aptamer, intramer, development; gene regulating aptamers, anti-TF aptamers for therapeutics, and conformation specific aptamers for allosteric inhibitors in cancers. However, in spite of great achievement of therapeutic intramers, the translation to the clinic is very slow. Hence, to facilitate the successful translation of therapeutic intramers, the few strategies have been considered.
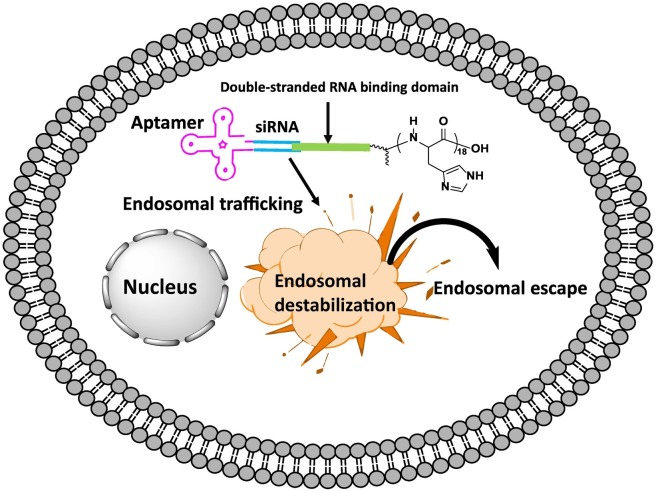
First, improvement of the endosomal escape. As described, internalized aptamers localize to endosomes and lysosomes. For biological activity, the aptamers must reach the cytosol, escaping the endosome. When the aptamers are endocytosed via a receptor-mediated clathrin-dependent mechanism, the rate of access to the cytosol mainly depends on the expression of receptors, the rates of receptor recycling, and endosomal escape efficiency. Typically, a small percentage (<0.01%) of endocytosed nucleic acid-based therapeutics are able to escape the endosome [122]. Hence, the facilitating of endosomal escape is pivotal to increase the efficacy of therapeutic intramers. For improving the endosomal escape, currently available endosomal escaping aids are small-molecule endosomolytic agents, such as chloroquine. An emerging alternative to improve endosomal escape is to conjugate endosomolytic peptides directly to the RNA molecules [122]. For example, anti-PMSA aptamer-siRNA chimeras were conjugated to pH-dependent polyhistidine (His) [123]. The His18 tag was shown to be remarkably effective in endosomal destabilization and improved the gene silencing effects of the chimera [123] (Fig. 5 ). Thus, conjugation of aptamers with tissue-penetrating or endosomal escaping peptides will facilitate the translation of therapeutic intramers to the clinic.
Fig. 5.
Endosomal escape of aptamers with peptides. Polyhistidine conjugation to an aptamer-siRNA chimera via double-stranded RNA binding domain (dsRBM) induces endosomal destabilization to enhance the release of the chimera into the cytosol.
Second, transcription-activating aptamers for therapeutics. Aptamer-mediated targeting of TFs is an emerging approach to suppress intractable protein targets. So far, transcription-activating aptamers mainly have been developed in E. coli, but not mammalian cells. Transcription activation and targeting of transcriptional cofactors with aptamers could lead to remarkable efficacy with surprisingly little toxicity to normal tissues. Therefore, membrane-bound transcription factors, such as Notch [124], will be also excellent targets for the development of therapeutic aptamers.
Third, Functional changes of truncated aptamers. Recently emerged major issue is the functional changes of aptamers after truncation. For instance, AMPA and kainite receptors are different subtypes of glutamate ion channels. One group found a full-length aptamer that blocked AMPA receptors specifically, but a truncated version of the same aptamer equally inhibited both the AMPA and kainite receptors. [125, 126], suggesting that the aptamer could serve multiple functions after truncation. Even though the dual function of this aptamer might be clinically beneficial, the functional or structural changes of aptamers after truncation should be carefully investigated.
Acknowledgments
Acknowledgements
We thank Dr. Kerin Higa at City of Hope for editorial assistance.
Funding
This work was supported by NIH grant AI029329 to J.J.R.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Aptamers in drug delivery”.
References
- 1.Que-Gewirth N.S., Sullenger B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 2.Gomes de Castro M.A., Hobartner C., Opazo F. Aptamers provide superior stainings of cellular receptors studied under super-resolution microscopy. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayor S., Pagano R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PubMed] [Google Scholar]
- 4.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 5.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 6.Liu A.P., Aguet F., Danuser G., Schmid S.L. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J. Cell Biol. 2010;191:1381–1393. doi: 10.1083/jcb.201008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Liu Z., Zou Y., Lai X., Ding D., Chen L., Zhang L., Wu Y., Chen Z., Tan W. Elucidating the cellular uptake mechanism of aptamer-functionalized graphene-isolated-Au-nanocrystals with dual-modal imaging. Analyst. 2016;141:3337–3342. doi: 10.1039/c6an00483k. [DOI] [PubMed] [Google Scholar]
- 8.Opazo F., Eiden L., Hansen L., Rohrbach F., Wengel J., Kjems J., Mayer G. Modular assembly of cell-targeting devices based on an uncommon G-quadruplex aptamer. Mol. Ther.–Nucleic Acids. 2015;4:e251. doi: 10.1038/mtna.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soundararajan S., Wang L., Sridharan V., Chen W., Courtenay-Luck N., Jones D., Spicer E.K., Fernandes D.J. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Hamhouyia F., Thomas S.D., Burke T.J., Girvan A.C., McGregor W.G., Trent J.O., Miller D.M., Bates P.J. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem. 2001;276:43221–43230. doi: 10.1074/jbc.M104446200. [DOI] [PubMed] [Google Scholar]
- 11.Reyes-Reyes E.M., Teng Y., Bates P.J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotula J.W., Pratico E.D., Ming X., Nakagawa O., Juliano R.L., Sullenger B.A. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012;22:187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranches G., Lukasser M., Schramek H., Ploner A., Stasyk T., Mayer G., Hüttenhofer A. In vitro selection of cell-internalizing DNA aptamers in a model system of inflammatory kidney disease. Mol. Ther.–Nucleic Acids. 2017;8:198–210. doi: 10.1016/j.omtn.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aptekar S., Arora M., Lawrence C.L., Lea R.W., Ashton K., Dawson T., Alder J.E., Shaw L. Selective targeting to glioma with nucleic acid aptamers. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L.Q., Xiao S.J., Hu P.P., Peng L., Ma J., Luo L.F., Li Y.F., Huang C.Z. Aptamer-mediated nanoparticle-based protein labeling platform for intracellular imaging and tracking endocytosis dynamics. Anal. Chem. 2012;84:3099–3110. doi: 10.1021/ac202810b. [DOI] [PubMed] [Google Scholar]
- 16.Cesur O., Nicol C., Groves H., Mankouri J., Blair G.E., Stonehouse N.J. The subcellular localisation of the human papillomavirus (HPV) 16 E7 protein in cervical cancer cells and its perturbation by RNA aptamers. Viruses. 2015;7:3443–3461. doi: 10.3390/v7072780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao H., Yang Z., Zhang S., Pang Z., Jiang X. Internalization and subcellular fate of aptamer and peptide dual-functioned nanoparticles. J. Drug Target. 2014;22:450–459. doi: 10.3109/1061186X.2014.886038. [DOI] [PubMed] [Google Scholar]
- 18.Liu C.H., Ren J., Liu C.M., Liu P.K. Intracellular gene transcription factor protein-guided MRI by DNA aptamers in vivo. FASEB J. 2014;28:464–473. doi: 10.1096/fj.13-234229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mongelard F., Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Bedford M.T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Teng Y., Girvan A.C., Casson L.K., Pierce W.M., Jr., Qian M., Thomas S.D., Bates P.J. AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin. Cancer Res. 2007;67:10491–10500. doi: 10.1158/0008-5472.CAN-06-4206. [DOI] [PubMed] [Google Scholar]
- 22.Paige J.S., Wu K.Y., Jaffrey S.R. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolgosheina E.V., Jeng S.C., Panchapakesan S.S., Cojocaru R., Chen P.S., Wilson P.D., Hawkins N., Wiggins P.A., Unrau P.J. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 25.Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strack R.L., Disney M.D., Jaffrey S.R. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Autour A., Westhof E., Ryckelynck M. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016;44:2491–2500. doi: 10.1093/nar/gkw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferre-D'Amare A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014;21:658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellenberger C.A., Wilson S.C., Sales-Lee J., Hammond M.C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013;135:4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponchon L., Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat. Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 32.Trachman R.J., III, Demeshkina N.A., Lau M.W.L., Panchapakesan S.S.S., Jeng S.C.Y., Unrau P.J., Ferre-D'Amare A.R. Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nat. Chem. Biol. 2017;13:807–813. doi: 10.1038/nchembio.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song W., Filonov G.S., Kim H., Hirsch M., Li X., Moon J.D., Jaffrey S.R. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol. 2017;13:1187–1194. doi: 10.1038/nchembio.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Autour A., S C.Y.J., A D.C., Abdolahzadeh A., Galli A., Panchapakesan S.S.S., Rueda D., Ryckelynck M., Unrau P.J. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018;9:656. doi: 10.1038/s41467-018-02993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeng S.C., Chan H.H., Booy E.P., McKenna S.A., Unrau P.J. Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers. RNA. 2016;22:1884–1892. doi: 10.1261/rna.056226.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilaratanakul V., Hauer D.A., Griffin D.E. Development and characterization of Sindbis virus with encoded fluorescent RNA aptamer Spinach2 for imaging of replication and immune-mediated changes in intracellular viral RNA. J. Gen. Virol. 2017;98:992–1003. doi: 10.1099/jgv.0.000755. [DOI] [PubMed] [Google Scholar]
- 37.Jepsen M.D.E., Sparvath S.M., Nielsen T.B., Langvad A.H., Grossi G., Gothelf K.V., Andersen E.S. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat. Commun. 2018;9:18. doi: 10.1038/s41467-017-02435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda M., Fourmy D., Yoshizawa S. Use of Baby Spinach and Broccoli for imaging of structured cellular RNAs. Nucleic Acids Res. 2017;45:1404–1415. doi: 10.1093/nar/gkw794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna-Rose W., Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 40.Latchman D.S. Inhibitory transcription factors. Int. J. Biochem. Cell Biol. 1996;28:965–974. doi: 10.1016/1357-2725(96)00039-8. [DOI] [PubMed] [Google Scholar]
- 41.Hunsicker A., Steber M., Mayer G., Meitert J., Klotzsche M., Blind M., Hillen W., Berens C., Suess B. An RNA aptamer that induces transcription. Chem. Biol. 2009;16:173–180. doi: 10.1016/j.chembiol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Ohuchi S., Suess B. An inhibitory RNA aptamer against the lambda cI repressor shows transcriptional activator activity in vivo. FEBS Lett. 2017;591:1429–1436. doi: 10.1002/1873-3468.12653. [DOI] [PubMed] [Google Scholar]
- 43.Sedlyarova N., Rescheneder P., Magan A., Popitsch N., Rziha N., Bilusic I., Epshtein V., Zimmermann B., Lybecker M., Sedlyarov V., Schroeder R., Nudler E. Natural RNA polymerase aptamers regulate transcription in E. coli. Mol. Cell. 2017;67:30–43. doi: 10.1016/j.molcel.2017.05.025. (e36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beilstein K., Wittmann A., Grez M., Suess B. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth. Biol. 2015;4:526–534. doi: 10.1021/sb500270h. [DOI] [PubMed] [Google Scholar]
- 45.Mondragon E., Maher L.J., III Anti-transcription factor RNA aptamers as potential therapeutics. Nucleic Acid Ther. 2016;26:29–43. doi: 10.1089/nat.2015.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raychaudhuri P., Park H.J. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halasi M., Gartel A.L. Targeting FOXM1 in cancer. Biochem. Pharmacol. 2013;85:644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 48.Xiang Q., Tan G., Jiang X., Wu K., Tan W., Tan Y. Suppression of FOXM1 transcriptional activities via a single-stranded DNA aptamer generated by SELEX. Sci. Rep. 2017;7 doi: 10.1038/srep45377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendillo M.L., Santagata S., Koeva M., Bell G.W., Hu R., Tamimi R.M., Fraenkel E., Ince T.A., Whitesell L., Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X., Shi H., Sevilimedu A., Liachko N., Nelson H.C., Lis J.T. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salamanca H.H., Antonyak M.A., Cerione R.A., Shi H., Lis J.T. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia Y., Shen S., Verma I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lebruska L.L., Maher L.J., III Selection and characterization of an RNA decoy for transcription factor NF-kappa B. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 55.Cassiday L.A., Maher L.J., III Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappa B. PNAS. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurster S.E., Maher L.J., III Selection and characterization of anti-NF-kappaB p65 RNA aptamers. RNA. 2008;14:1037–1047. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Speck N.A. Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol. 2001;8:192–196. doi: 10.1097/00062752-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 58.Mikhail F.M., Sinha K.K., Saunthararajah Y., Nucifora G. Normal and transforming functions of RUNX1: a perspective. J. Cell. Physiol. 2006;207:582–593. doi: 10.1002/jcp.20538. [DOI] [PubMed] [Google Scholar]
- 59.Barton J.L., Bunka D.H., Knowling S.E., Lefevre P., Warren A.J., Bonifer C., Stockley P.G. Characterization of RNA aptamers that disrupt the RUNX1-CBFbeta/DNA complex. Nucleic Acids Res. 2009;37:6818–6830. doi: 10.1093/nar/gkp728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto R., Katahira M., Nishikawa S., Baba T., Taira K., Kumar P.K. A novel RNA motif that binds efficiently and specifically to the Ttat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells. 2000;5:371–388. doi: 10.1046/j.1365-2443.2000.00330.x. [DOI] [PubMed] [Google Scholar]
- 62.Tuerk C., MacDougal S., Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. PNAS. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukuda K., Vishnuvardhan D., Sekiya S., Hwang J., Kakiuchi N., Taira K., Shimotohno K., Kumar P.K., Nishikawa S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000;267:3685–3694. doi: 10.1046/j.1432-1327.2000.01400.x. [DOI] [PubMed] [Google Scholar]
- 64.Hwang B., Cho J.S., Yeo H.J., Kim J.H., Chung K.M., Han K., Jang S.K., Lee S.W. Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA. 2004;10:1277–1290. doi: 10.1261/rna.7100904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biroccio A., Hamm J., Incitti I., De Francesco R., Tomei L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002;76:3688–3696. doi: 10.1128/JVI.76.8.3688-3696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.S.A.o.P.T.b.B German advisory committee blood, human immunodeficiency virus (HIV) Transfus. Med. Hemother. 2016;43:203–222. doi: 10.1159/000445852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanchez-Luque F.J., Stich M., Manrubia S., Briones C., Berzal-Herranz A. Efficient HIV-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014;4:6242. doi: 10.1038/srep06242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P., Wilson S.M., King R.W., Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J.H., Shin S.K., Jiang Y., Choi W.H., Hong C., Kim D.E., Lee M.J. Facilitated tau degradation by USP14 aptamers via enhanced proteasome activity. Sci. Rep. 2015;5 doi: 10.1038/srep10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gillan L., Matei D., Fishman D.A., Gerbin C.S., Karlan B.Y., Chang D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 73.Roy S., Patel D., Khanna S., Gordillo G.M., Biswas S., Friedman A., Sen C.K. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. PNAS. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kudo Y., Siriwardena B.S., Hatano H., Ogawa I., Takata T. Periostin: novel diagnostic and therapeutic target for cancer. Histol. Histopathol. 2007;22:1167–1174. doi: 10.14670/HH-22.1167. [DOI] [PubMed] [Google Scholar]
- 75.Lee Y.J., Kim I.S., Park S.A., Kim Y., Lee J.E., Noh D.Y., Kim K.T., Ryu S.H., Suh P.G. Periostin-binding DNA aptamer inhibits breast cancer growth and metastasis. Mol. Ther. 2013;21:1004–1013. doi: 10.1038/mt.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brosius F.C., Khoury C.C., Buller C.L., Chen S. Abnormalities in signaling pathways in diabetic nephropathy. Expert. Rev. Endocrinol. Metab. 2010;5:51–64. doi: 10.1586/eem.09.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma K., Ziyadeh F.N. Renal hypertrophy is associated with upregulation of TGF-beta 1 gene expression in diabetic BB rat and NOD mouse. Am. J. Phys. 1994;267 doi: 10.1152/ajprenal.1994.267.6.F1094. (F1094-1001) [DOI] [PubMed] [Google Scholar]
- 78.Lan H.Y. Transforming growth factor-beta/Smad signalling in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 2012;39:731–738. doi: 10.1111/j.1440-1681.2011.05663.x. [DOI] [PubMed] [Google Scholar]
- 79.Horiuchi K., Amizuka N., Takeshita S., Takamatsu H., Katsuura M., Ozawa H., Toyama Y., Bonewald L.F., Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J. Bone Miner. Res. 1999;14:1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 80.Um J.E., Park J.T., Nam B.Y., Lee J.P., Jung J.H., Kim Y., Kim S., Park J., Wu M., Han S.H., Yoo T.H., Kang S.W. Periostin-binding DNA aptamer treatment attenuates renal fibrosis under diabetic conditions. Sci. Rep. 2017;7:8490. doi: 10.1038/s41598-017-09238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sugiyama A., Kanno K., Nishimichi N., Ohta S., Ono J., Conway S.J., Izuhara K., Yokosaki Y., Tazuma S. Periostin promotes hepatic fibrosis in mice by modulating hepatic stellate cell activation via alphav integrin interaction. J. Gastroenterol. 2016;51:1161–1174. doi: 10.1007/s00535-016-1206-0. [DOI] [PubMed] [Google Scholar]
- 82.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29:366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shigdar S., Qiao L., Zhou S.F., Xiang D., Wang T., Li Y., Lim L.Y., Kong L., Li L., Duan W. RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 2013;330:84–95. doi: 10.1016/j.canlet.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 84.Zhou G., Latchoumanin O., Bagdesar M., Hebbard L., Duan W., Liddle C., George J., Qiao L. Aptamer-based therapeutic approaches to target cancer stem cells. Theranostics. 2017;7:3948–3961. doi: 10.7150/thno.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van de Pavert S.A., Meuleman J., Malysheva A., Aartsen W.M., Versteeg I., Tonagel F., Kamphuis W., McCabe C.J., Seeliger M.W., Wijnholds J. A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J. Neurosci. 2007;27:564–573. doi: 10.1523/JNEUROSCI.3496-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hrascan R., Pavelic K., Pavicic D., Krizanac S., Stajcer-Sittic V., Pecur L., Spaventi S., Klimpfinger M., Pavelic J. Concomitant point mutation of tumor suppressor gene p53 and oncogene c-N-ras in malignant neuroendocrine pancreatic tumor. Anticancer Res. 1996;16:3761–3766. [PubMed] [Google Scholar]
- 87.Bullock A.N., Fersht A.R. Rescuing the function of mutant p53. Nat. Rev. Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 88.Blandino G., Levine A.J., Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 89.Chen L., Rashid F., Shah A., Awan H.M., Wu M., Liu A., Wang J., Zhu T., Luo Z., Shan G. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. PNAS. 2015;112:10002–10007. doi: 10.1073/pnas.1502159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kovacs J.J., Hara M.R., Davenport C.L., Kim J., Lefkowitz R.J. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev. Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Srivastava A., Gupta B., Gupta C., Shukla A.K. Emerging functional divergence of beta-arrestin isoforms in GPCR function. Trends Endocrinol. Metab. 2015;26:628–642. doi: 10.1016/j.tem.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 92.Fereshteha M., Ito T., Kovacs J.J., Zhao C., Kwon H.Y., Tornini V., Konuma T., Chen M.Y., Lefkowitz R.J., Reya T. beta-Arrestin2 mediates the initiation and progression of myeloid leukemia. PNAS. 2012;109:12532–12537. doi: 10.1073/pnas.1209815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kotula J.W., Sun J., Li M., Pratico E.D., Fereshteh M.P., Ahrens D.P., Sullenger B.A., Kovacs J.J. Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 95.Pelletier J., Graff J., Ruggero D., Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raza F., Waldron J.A., Quesne J.L. Translational dysregulation in cancer: eIF4A isoforms and sequence determinants of eIF4A dependence. Biochem. Soc. Trans. 2015;43:1227–1233. doi: 10.1042/BST20150163. [DOI] [PubMed] [Google Scholar]
- 97.Modelska A., Turro E., Russell R., Beaton J., Sbarrato T., Spriggs K., Miller J., Graf S., Provenzano E., Blows F., Pharoah P., Caldas C., Le Quesne J. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oguro A., Ohtsu T., Svitkin Y.V., Sonenberg N., Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsieh A.C., Costa M., Zollo O., Davis C., Feldman M.E., Testa J.R., Meyuhas O., Shokat K.M., Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Famulok M., Mayer G. Intramers and aptamers: applications in protein-function analyses and potential for drug screening. Chembiochem. 2005;6:19–26. doi: 10.1002/cbic.200400299. [DOI] [PubMed] [Google Scholar]
- 101.Lagerstrom M.C., Schioth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 102.Kahsai A.W., Xiao K., Rajagopal S., Ahn S., Shukla A.K., Sun J., Oas T.G., Lefkowitz R.J. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat. Chem. Biol. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manglik A., Kim T.H., Masureel M., Altenbach C., Yang Z., Hilger D., Lerch M.T., Kobilka T.S., Thian F.S., Hubbell W.L., Prosser R.S., Kobilka B.K. Structural insights into the dynamic process of beta2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kahsai A.W., Wisler J.W., Lee J., Ahn S., Cahill T.J., III, Dennison S.M., Staus D.P., Thomsen A.R., Anasti K.M., Pani B., Wingler L.M., Desai H., Bompiani K.M., Strachan R.T., Qin X., Alam S.M., Sullenger B.A., Lefkowitz R.J. Conformationally selective RNA aptamers allosterically modulate the beta2-adrenoceptor. Nat. Chem. Biol. 2016;12:709–716. doi: 10.1038/nchembio.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swain J.F., Dinler G., Sivendran R., Montgomery D.L., Stotz M., Gierasch L.M. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thirunavukarasu D., Shi H. An RNA aptamer specific to Hsp70-ATP conformation inhibits its ATPase activity independent of Hsp40. Nucleic Acid Ther. 2015;25:103–112. doi: 10.1089/nat.2014.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karin M., Liu Z., Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 108.Wisniewska M.B., Ameyar-Zazoua M., Bakiri L., Kaminska B., Yaniv M., Weitzman J.B. Dimer composition and promoter context contribute to functional cooperation between AP-1 and NFAT. J. Mol. Biol. 2007;371:569–576. doi: 10.1016/j.jmb.2007.05.079. [DOI] [PubMed] [Google Scholar]
- 109.Eferl R., Wagner E.F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 110.Walters R.D., McSwiggen D.T., Goodrich J.A., Kugel J.F. Selection and characterization of a DNA aptamer that can discriminate between cJun/cJun and cJun/cFos. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Walters R.D., Drullinger L.F., Kugel J.F., Goodrich J.A. NFATc2 recruits cJun homodimers to an NFAT site to synergistically activate interleukin-2 transcription. Mol. Immunol. 2013;56:48–56. doi: 10.1016/j.molimm.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fay M.M., Lyons S.M., Ivanov P. RNA G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017;429:2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Magbanua E., Zivkovic T., Hansen B., Beschorner N., Meyer C., Lorenzen I., Grotzinger J., Hauber J., Torda A.E., Mayer G., Rose-John S., Hahn U. d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 2013;10:216–227. doi: 10.4161/rna.22951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Esposito V., Pirone L., Mayol L., Pedone E., Virgilio A., Galeone A. Exploring the binding of d(GGGT)4 to the HIV-1 integrase: an approach to investigate G-quadruplex aptamer/target protein interactions. Biochimie. 2016;127:19–22. doi: 10.1016/j.biochi.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 115.Saito T., Yoshida W., Yokoyama T., Abe K., Ikebukuro K. Identification of RNA oligonucleotides binding to several proteins from potential G-quadruplex forming regions in transcribed pre-mRNA. Molecules. 2015;20:20832–20840. doi: 10.3390/molecules201119733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Avci-Adali M., Steinle H., Michel T., Schlensak C., Wendel H.P. Potential capacity of aptamers to trigger immune activation in human blood. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dunn M.R., Jimenez R.M., Chaput J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1 [Google Scholar]
- 118.Yoon S., Rossi J.J. Future strategies for the discovery of therapeutic aptamers. Expert Opin. Drug Discovery. 2017;12:317–319. doi: 10.1080/17460441.2017.1290077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee Y., Urban J.H., Xu L., Sullenger B.A., Lee J. 2'Fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Ther. 2016;26:173–182. doi: 10.1089/nat.2015.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song K.M., Lee S., Ban C. Aptamers and their biological applications. Sensors. 2012;12:612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ganson N.J., Povsic T.J., Sullenger B.A., Alexander J.H., Zelenkofske S.L., Sailstad J.M., Rusconi C.P., Hershfield M.S. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J. Allergy Clin. Immunol. 2016;137:1610–1613. doi: 10.1016/j.jaci.2015.10.034. (e1617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 123.Liu H.Y., Gao X. A universal protein tag for delivery of SiRNA-aptamer chimeras. Sci. Rep. 2013;3:3129. doi: 10.1038/srep03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kopan R. Notch: a membrane-bound transcription factor. J. Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 125.Jaremko W.J., Huang Z., Wen W., Wu A., Karl N., Niu L. Identification and characterization of RNA aptamers: a long aptamer blocks the AMPA receptor and a short aptamer blocks both AMPA and kainate receptors. J. Biol. Chem. 2017;292:7338–7347. doi: 10.1074/jbc.M116.774752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jaremko W.J., Huang Z., Wen W., Wu A., Karl N., Niu L. One aptamer, two functions: the full-length aptamer inhibits AMPA receptors, while the short one inhibits both AMPA and kainate receptors. RNA Dis. 2017;4 [PMC free article] [PubMed] [Google Scholar]