Figure 5.
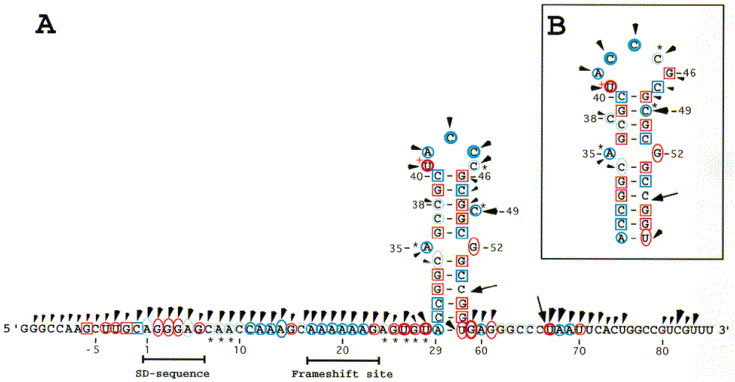
Imidazole induced cleavages and mapping data of Watson-Crick positions of the dnaX frameshift region RNA. Imidazole cleavages are shown on the RNA sequence with arrowheads. The size of the arrowheads refers to the intensity of cleavages (weak, intermediate, and strong). Chemical reactivity of the Watson-Crick positions are indicated on the RNA sequence. DMS reactivities are indicated in blue and CMCT reactivities in red. Reactivities under native conditions (○), under semi-denaturing conditions but not in native conditions (0), or no reactivities under both native and semi-denaturing conditions (□) of adenine residues (atom N-1), cytosine residues (atom N-3), guanine residues (atom N-1), and uracil residues (atom N-3). Weak, intermediate, and strong reactivities are denoted by light, thin, and thick symbols. (∗) indicates that the Watson-Crick position is more strongly modified under semi-denaturing conditions than under native conditions. The two arrows indicate degradation seen in the control lanes. A very strong CMCT modification is denoted by a+next to the nucleotide (U41).
