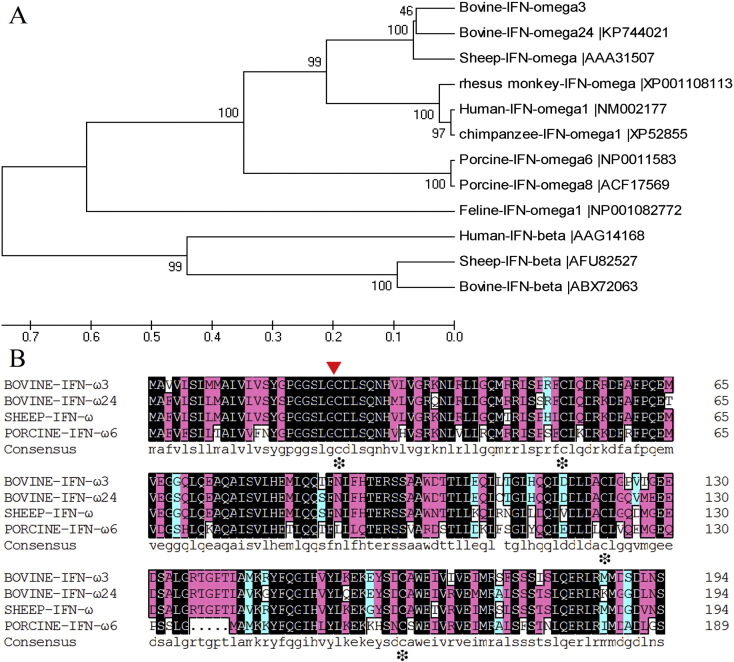
Fig. 2.
Phylogenetic tree construction of the BoIFN-ω3 and IFN-ω amino acid alignment. (A) The phylogenetic tree was made with MEGA 5.0 using the UPGMA method, while the availability of the branch stem was verified with the Bootstrap using 500 parameters. The scale bar represents the genetic distance. The credibility value for each node is shown. (B) IFN-ω amino acid alignment of bovine, sheep, and porcine. The arrow denotes the signal sequence cleavage site. Identical residues are in black boxes, 75%-conserved residues are in pink boxes, and semi-conserved residues were in light blue boxes. White boxes are amino acids that differ between the sequences. The conserved cysteine residues are marked with (*).