Figure I.
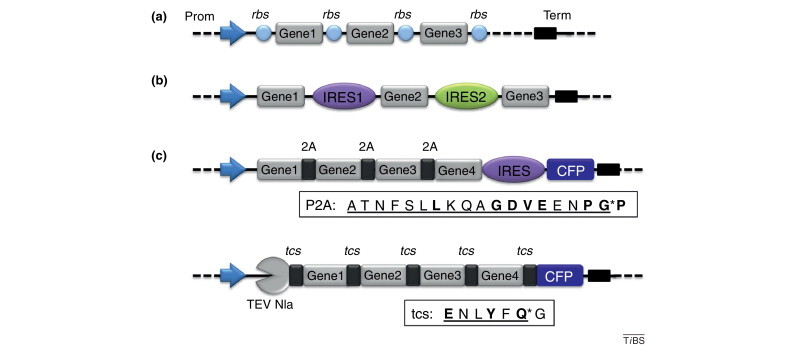
Tools for expression of protein complexes. (a) Protein complexes can be produced efficiently in E. coli by using polycistrons. In polycistrons, genes of interest are spaced apart by ribosome binding sites (rbs) and placed under control of a single promoter (Prom). A sequence (Term) ensures termination of mRNA transcription. One or several polycistrons can be provided on one or more plasmids simultaneously in one cell. (b) The equivalent construction for eukaryotic expression provides IRESs instead of rbs, resulting also in polycistronic mRNAs. (c) Polyproteins are particularly useful to balance the stoichiometry of expressed proteins. Polyprotein constructions can provide self-cleaving peptides derived from picornavirus (2A) in between the individual proteins (top). A fluorescent protein (here CFP) can be included to facilitate tracking of heterologous expression from the respective promoter by fluorescence measurement. As an alternative, polyproteins can be used that contain a highly specific protease [here tobacco etch virus (TEV) Nla], which processes the polyprotein by cleaving corresponding cleavage sites (tcs), thus liberating the individual proteins including itself (bottom). Both polyprotein strategies have the drawback of decorating the C terminus of the preceding protein with overhanging residues that are part of the cleavage sequence (boxed). Asterisks denote sites of cleavage. Conserved residues defining the site are in bold. C-terminal overhangs are underlined.
