Fig. 2.
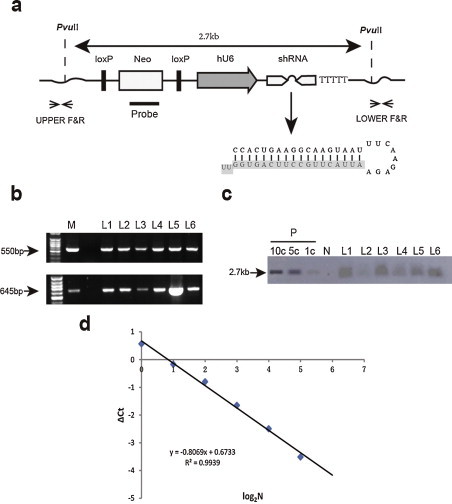
Analysis of transgene integration. (a) Schematic diagram of transgenic vector (pshRNA-1B372). The head-to-head arrows denote the primers used to identify the transgenic insert in TG pigs. Two PvuII sites, 2.7 kb apart, and the probe position used for Southern blotting are indicated. (b) PCR for detecting TG mice and assessment of integrity of the inserts. The two ends of pshRNA-1B372 transgenes were amplified (550 bp and 645 bp) separately from TG pig genomic DNA using 5′- and 3′-specific primers. Two PCR products at the length of 550 bp and 645 bp were identified as 5′ end and 3′ end of the inserted transgene. M, 100 bp DNA ladder; L1–L6, TG founders; N, genomic DNA from WT pig as a negative control; P, pshRNA-1B372 as a positive control. (c) Detection of the transgene integrated into the genome by Southern blotting analysis. The siRNA expression cassette was inserted into the genome, as indicated by the band. The positive control was the vector plasmid. After electrophoresis and Southern blot transfer, membranes were probed for integrated vector DNA. Copy number standards (the number of inserted vector DNA per diploid genomic equivalent) are indicated as 1c, 3c and 5c. (d) A standard curve for the number of transgenes and the corresponding Ct value. QRT-PCR was performed to determine the copy number of transgene. A standard set of mixtures, including 1, 2, 4, 8, 16, and 32 copies of plasmid DNA in 10 ng of WT pig genomic DNA, was used to establish a standard curve. The Y axis represents the ΔCt value (Ct of pshRNA-1b372 gene minus Ct of MSTN) and X axis represents the log2 ratio of the transgene copy number.
