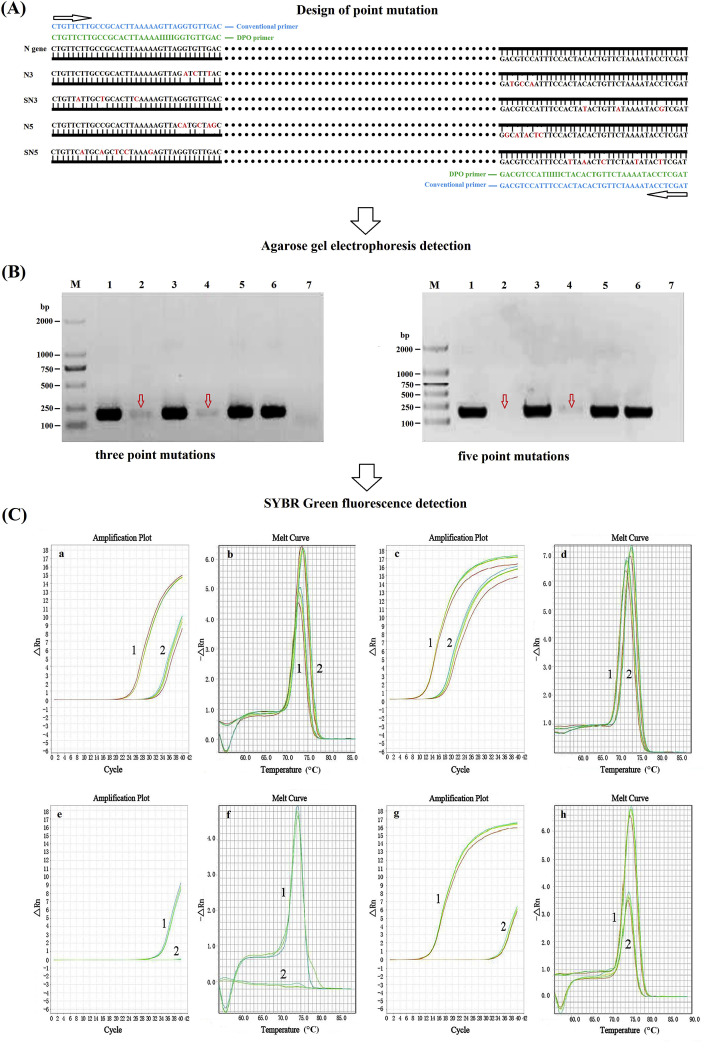
Fig. 1.
Comparison of priming specificity between DPO primer and conventional primer. A: Point mutations targeting gene sequences of the 5′- or 3′-segments of DPO primer (3′-segment with three/five sites mutation namely N3/N5; 5′-segment with three/five sites mutation namely SN3/SN5), followed by PCR amplification using conventional PCR assay and SYBR Green-based real-time PCR assay with conventional primer (CP) and DPO primer, respectively. B: PCR products were analyzed by agarose gel electrophoresis. (Left) M: DNA Marker DL2000; 1: N3+CP; 2: N3+DPO; 3: SN3+CP; 4: SN3+DPO; 5: N gene + CP; 6: N gene + DPO; 7: Negative control. (Right) M: DNA Marker DL2000; 1: N5+CP; 2: N5+DPO; 3: SN5+CP; 4: SN5+DPO; 5: N gene + CP; 6: N gene + DPO; 7: Negative control. C: SYBR Green fluorescence detection results. Detection result of three nucleotide mismatches (a/c: amplification plot; b/d: melt curve). 1: N3/SN3+ conventional primer; 2: N3/SN3+DPO. Detection result of five nucleotide mismatches (e/g: amplification plot; f/h: melt curve). 1: N5/SN5+CP; 2: N5/SN5+DPO.