Fig. 3.
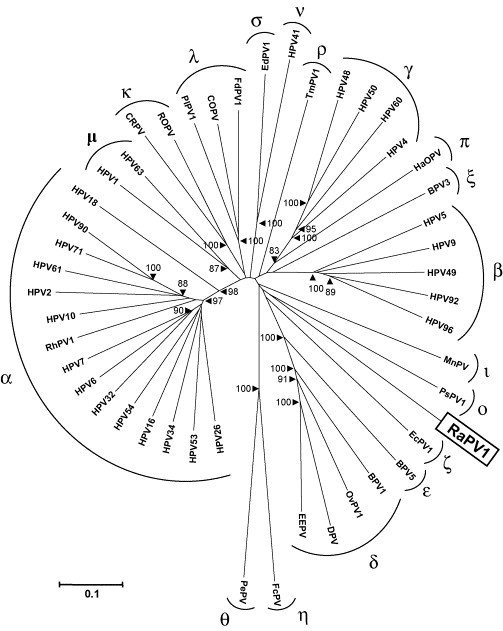
Neighbor-joining phylogenetic tree, based on a concatenated E1/E2/L2/L1 nucleotide sequence alignment of RaPV-1 and 46 other PV type species of the different PV genera and species. The PV-types included (with their GenBank accession numbers) were bovine BPV-1 (NC_001522), BPV-3 (NC_004197), BPV-5 (NC_004195), canine oral COPV (NC_001619), cottontail rabbit CRPV (NC_001541), deer DPV (NC_001523), Equus caballus EcPV-1 (NC_003748), Erethizon dorsatum EdPV-1 (NC_006951) European elk EEPV (NC_001524), Fringilla coelebs FcPV (NC_004068), Felis domesticus FdPV-1 (NC_004765), hamster oral HaOPV (E15111), HPV-1a (NC_001356), HPV-2a (NC_001352), HPV-4 (NC_001457), HPV-5 (NC_001531), HPV-6b (NC_001355), HPV-7 (NC_001595), HPV-9 (NC_001596), HPV-10 (NC_001576), HPV-16 (NC_001526), HPV-18 (NC_001357), HPV-26 (NC_001583), HPV-32 (NC_001586), HPV-34 (NC_001587), HPV-41 (NC_001354), HPV-48 (NC_001690), HPV-49 (NC_001591), HPV-50 (NC_001691), HPV-53 (NC_001593), HPV-54 (NC_001676), HPV-60 (NC_001693), HPV-61 (NC_001694), HPV-63 (NC_001458), HPV-71 (NC_002644), HPV-90 (NC_004104), HPV-92 (NC_004500), HPV-96 (NC_005134), Mastomys natalensis MnPV (NC_001605), ovine OvPV-1 (NC_001789), Psittacus erithacus PePV (NC_003973), Procyon lotor PlPV-1 (AY763115), Phocoena spinipinnis PsPV-1 (NC_003348), Rousettus aegyptiacus RaPV-1 (DQ366842), rhesus monkey RhPV-1 (NC_001678), rabbit oral ROPV (NC_002232), Trichechus manatus latirostris TmPV-1 (NC_006563). The PV genera are indicated with their Greek symbols. The numbers at the internal nodes represent the bootstrap probabilities (in percent), as determined for 1000 iterations by the neighbor-joining method. Only bootstrap values greater than 80% are shown. The scale bar indicates the genetic distance (nucleotide substitutions per site).
