Figure 1.
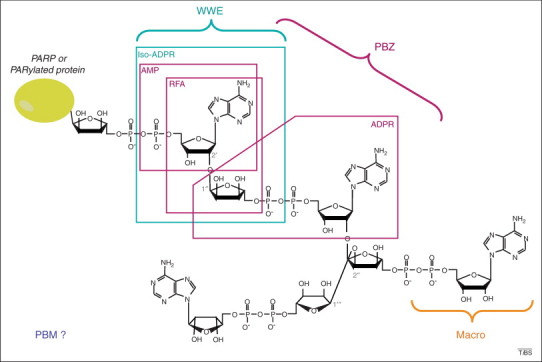
Structure of poly(ADP-ribose) (PAR) and recognition by PAR-binding motifs (PBMs). The chemical structure of branched PAR, covalently attached to an acceptor protein (which can be poly(ADP-ribose) polymerase itself), is shown. The characteristic linkages of linear (1→2′ O-glycosidic bound) and branched (1→2′′ O-glycosidic bound) are indicated. The brackets illustrate the region recognized by each PAR-binding domain. The ADP-ribose (ADPR) derivatives used in structural characterization of PAR-binding domains are framed. The RNF146 PAR-binding domain WWE was co-crystallized with iso-ADPR [46], supporting a mechanism of internal recognition of PAR. The solution structure of the two aprataxin and PNK-like factor (APLF) PAR-binding zinc fingers (PBZ motifs) bound to 2′-O-α-D-ribofuranosyl-adenosine (RFA) was solved by nuclear magnetic resonance (NMR) [38], whereas the CHFR PBZ domain was co-crystallized with AMP and ADPR [41]. This PBZ domain was also co-crystallized with P1P2-diadenosine 5′-pyrophosphate (AMP2), which is not a derivative of PAR and thus cannot be illustrated in this scheme, but which supports the hypothesis that the PBZ motif recognizes two successive ADPR units. Macrodomains bind ADPR units, but capping of the 2′OH from ADP prohibits internal recognition of PAR 14, 24. How the PBMs recognize PAR remains unknown, as illustrated by the open question mark.
