Fig. 4.
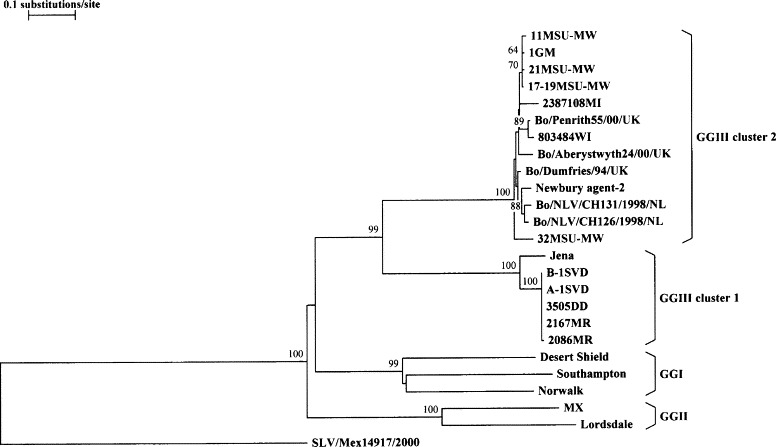
Phylogenetic analysis based on deduced amino acid sequences corresponding to a 744-nucleotide central variable region of the capsid gene of bovine noroviruses 11MSU-MW, 1GM, 21MSU-MW, 17–19MSU-MW, 2387108MI, 803484WI, 32MSU-MW, B-1SVD, A-1SVD, 3505DD, 2167MR, and 2086MR (this study); Jena (AJ011099); Bo/Penrith55/00/UK (AY126476); Bo/Aberystwyth24/00/UK (AY126475); Bo/Dumfries/94/UK (AY126474); Newbury agent-2 (AF097917); Bo/NLV/CH131/1998/NL (AF320113); Bo/NLV/CH126/1998/NL (AF320625); and of human noroviruses Desert Shield (U04469), Southampton (L07418), Norwalk (M87661), MX (U22498) and Lordsdale (X86557). Human sapovirus strain, SLV/Mex14917/2000 (AF435813), was included as the outgroup sequence. GenBank accession numbers are in parentheses. Bootstrap values at nodes are given if >60%.
