Fig. 4.
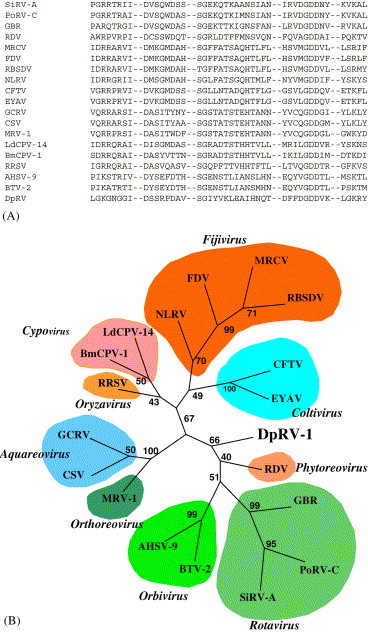
Phylogenetic analysis of the RNA-dependent-RNA polymerase (RdRp) of DpRV-1. (A) Alignment of the conserved motifs of the RdRp of reoviruses. The sequences used were from the following viruses: from of the genus Rotavirus; Simian rotavirus SA11 (SirV-A; P22678), Porcine rotavirus (PoRV-C; M74216), Group B rotavirus (GBR; P35942); from the genus Phytoreovirus; Rice dwarf virus (RDV; U73201); From the genus Fijivirus; Mal de Rio Cuarto virus (MRCV; AF49925), Fiji disease virus (FDV; AY029520), Rice black streaked dwarf virus (RBSDV; AJ294757), Nilaparvata lugens reovirus (NLRV; D49693); from the genus Coltivirus; Colorado tick fever virus (CFTV; AF133428), Eyach virus (EYAV; AF282467); from the genus Aquareovirus; Grass carp reovirus (GCRV; AF260511S2), Chulm salmon reovirus (CSV; AF418295); from the genus Orthoreovirus; Mammalian orthoreovirus subgroup 1 (MRV-1; M24734); from the genus Cypovirus; Bombyx mori cypovirus (BmCPV-1; AF323782), Lymantria dispar cypovirus (LdCPV-14; AF389452); from the genus Oryzavirus; Rice ragged stunt virus (RRSV; U66714), from the genus Orbivirus; African horse sickness virus (ASFV-9; U944887), Blue tongue virus (BTV-2; L20508) and from the unassigned reovirus, DpRV-1. (B) Phylogenetic unrooted tree of RdRp obtained by the Parsimony method. The same topology was obtained using the Neighbor-joining method.
