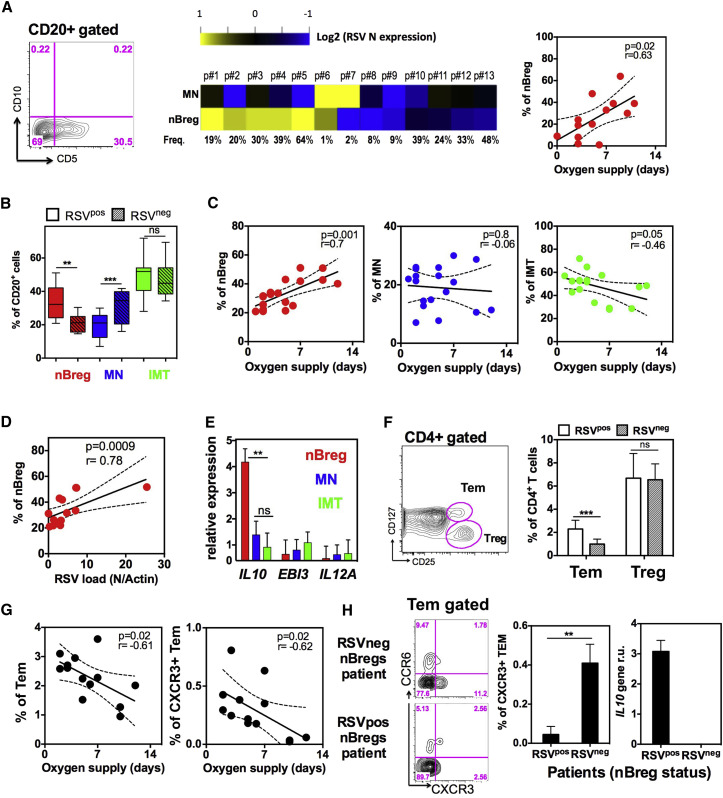
Figure 7.
nBreg Cells Are Infected by RSV in Patients and Predict the Severity of Acute Bronchiolitis
(A) CD20+ B cells in nasopharyngeal aspirates (NPA) collected from RSV-positive patients (n = 13) at the first day of hospital admission were analyzed and purified by FACS as nBreg cells and MN B cell subsets based on CD5 and CD10 expression. Heatmap shows sorted cell subsets analyzed for RSV nucleoprotein gene (RSV N) expression by qRT-PCR. Correlation plots (right) of nBreg cell frequency in the NPA with the duration of oxygen supply.
(B) Frequency of B cells subsets in the blood of RSV-positive (RSVpos; n = 18) and -negative (RSVneg; n = 10) patients.
(C) Correlation plot of B cell subset percentage in the blood of RSVpos patients (n = 18) with the duration of oxygen supply.
(D) Correlation plot of blood nBreg cell frequency with RSV load in the corresponding NPA of RSVpos patients (n = 13).
(E) nBreg, MN, and IMT B cells were sorted from the blood of RSVpos patients (n = 7) and stimulated or not with HRSV for 24 hr. IL10, EBI3, and IL12 gene expression was analyzed by qRT-PCR and normalized to housekeeping genes.
(F) CD4+ T cells were analyzed by FACS for Treg and Tem cells, as shown in the blood of RSV-positive and (RSVpos; n = 13) and -negative (RSVneg; n = 7) patients.
(G) Correlation plots of blood Tem and CXCR3+ Tem cell frequencies with the duration of oxygen supplementation (n = 13).
(H) Representative FACS analysis plot of blood CXCR3+ and CCR6+ Tem cells in patients whose nBreg cells were found infected or not in the NPA of (A). Histograms represent the frequency of CXCR3+ Tem cells in the blood (left histogram) and nBreg cells IL-10 gene expression in the NPA (right histogram) from patients corresponding to infected nBreg (RSVpos; n = 3) or non-infected nBreg (RSVneg; n = 3) cells.
Results are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ns for non significant.