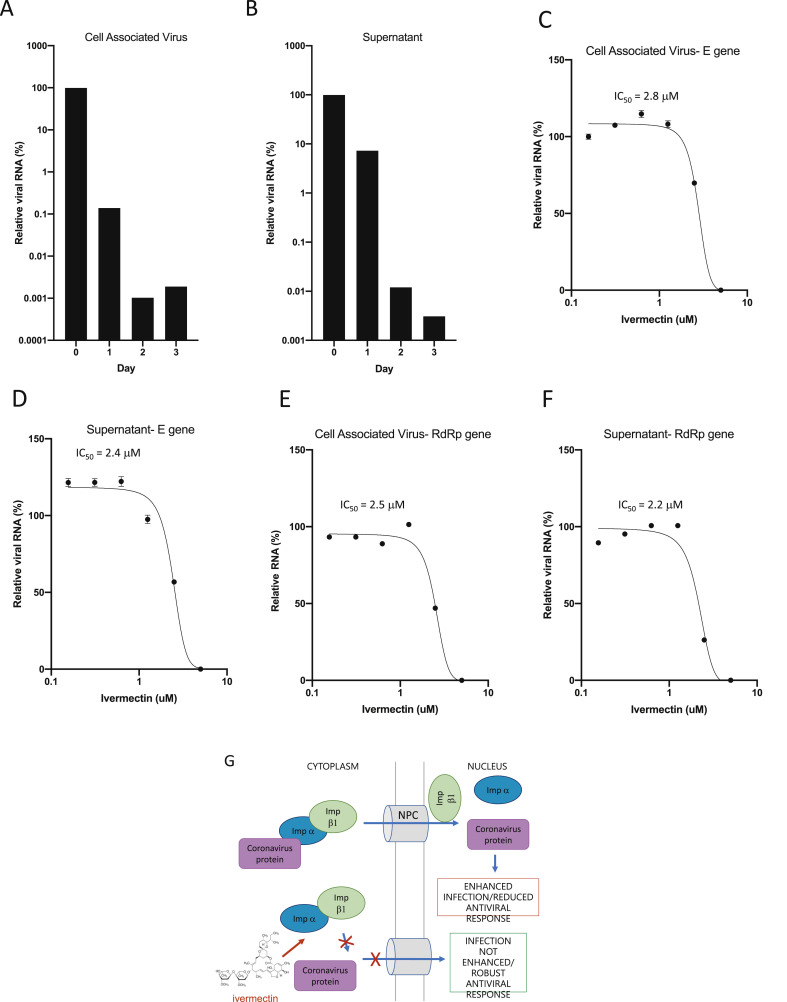
Fig. 1.
Ivermectin is a potent inhibitor of the SARS-CoV-2 clinical isolate Australia/VIC01/2020. Vero/hSLAM cells were in infected with SARS-CoV-2 clinical isolate Australia/VIC01/2020 (MOI = 0.1) for 2 h prior to addition of vehicle (DMSO) or Ivermectin at the indicated concentrations. Samples were taken at 0–3 days post infection for quantitation of viral load using real-time PCR of cell associated virus (A) or supernatant (B). IC50 values were determined in subsequent experiments at 48 h post infection using the indicated concentrations of Ivermectin (treated at 2 h post infection as per A/B). Triplicate real-time PCR analysis was performed on cell associated virus (C/E) or supernatant (D/F) using probes against either the SARS-CoV-2 E (C/D) or RdRp (E/F) genes. Results represent mean ± SD (n = 3). 3 parameter dose response curves were fitted using GraphPad prism to determine IC50 values (indicated). G. Schematic of ivermectin's proposed antiviral action on coronavirus. IMPα/β1 binds to the coronavirus cargo protein in the cytoplasm (top) and translocates it through the nuclear pore complex (NPC) into the nucleus where the complex falls apart and the viral cargo can reduce the host cell's antiviral response, leading to enhanced infection. Ivermectin binds to and destabilises the Impα/β1 heterodimer thereby preventing Impα/β1 from binding to the viral protein (bottom) and preventing it from entering the nucleus. This likely results in reduced inhibition of the antiviral responses, leading to a normal, more efficient antiviral response.