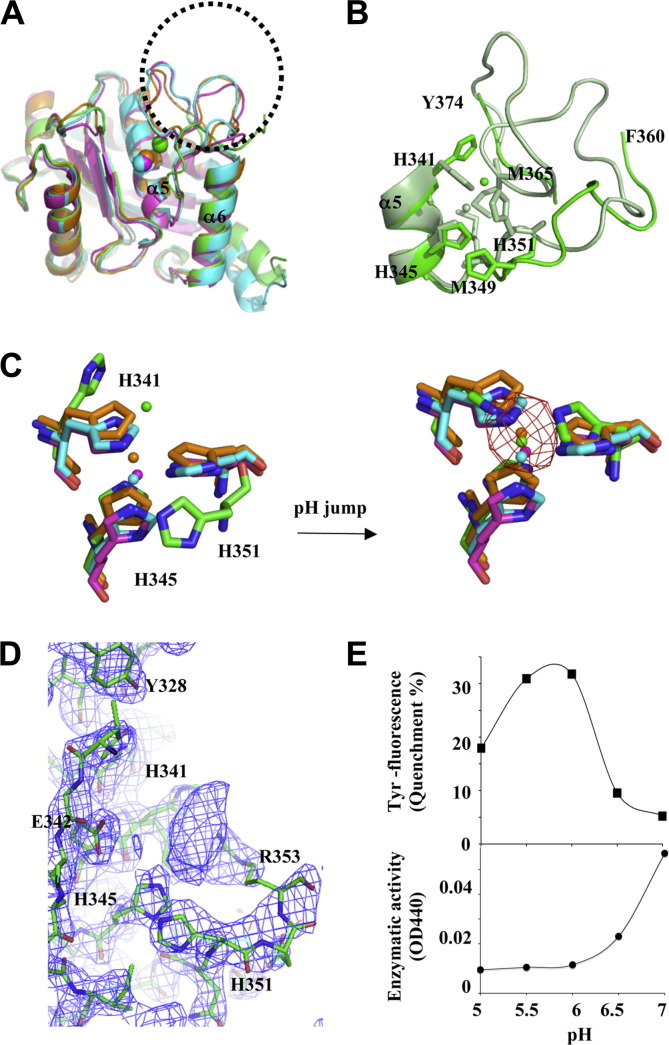
Fig. 2.
The pH-dependent zinc-binding motif on the metalloprotease domain and the associated proteolytic activity. (A) Superimposition of M domains of four P-III SVMPs: atragin (green), K-like (orange), VAP1 (cyan), and VAP2 (pink). The major variable region is located at the loop containing the Met-turn (enclosed by the dotted circle), which is disordered in the atragin structure. (B) The atragin structure in the acidic crystal (pH 5.0) (green) reveals an unconventional zinc-binding motif resulting in the missing loop from Asn361 to Ala373, whereas in a neutral crystal (pH 7.4) (light green), the binding motif switches to the conventional state and the electron density of the missing loop appears to allow model building. (C) Superimposition of three histidine residues in zinc-binding motifs of four P-III SVMPs shows the unconventional zinc-binding motif of atragin (green) in acidic crystallization conditions (citric acid, pH 5.0) and the conventional motif of atragin in the soaked neutral condition (Hepes, pH 7.4). The zinc anomalous difference-Fourier map (cutoff = 5σ, red mesh) confirms the zinc location in the neutral condition but not in the acidic condition. (D) The composite omit |2Fo − Fc| map (cutoff = 1.5σ, blue mesh) shows the well fit of three histidine residues in an unconventional binding motif and the putative range of the zinc position which is unobservable in the anomalous difference-Fourier map in the acidic pH crystal. The side chain of His341 has two conformations, and the backbone of His351 is repelled compared with a conventional zinc-binding motif. (E) The pH effect on the Tyr fluorescence and enzymatic activity of atragin. At pH 5.0 (acidic crystallization buffer), the enzymatic activity decreased to ∼20%, and the Tyr fluorescence also decreased ∼20% relative to assays at the neutral pH (pH 7.4). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)