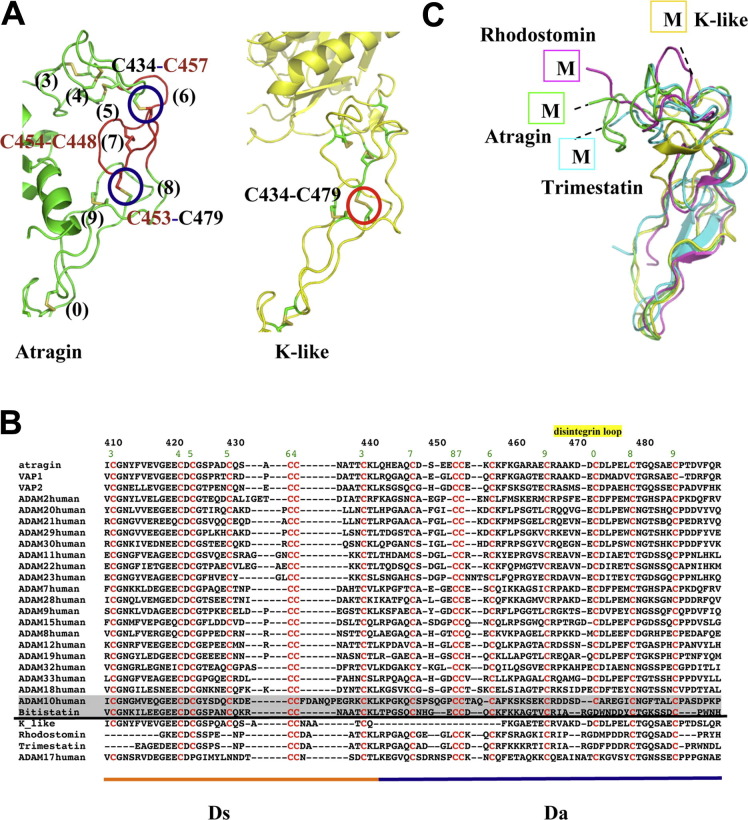
Fig. 4.
Disulfide bond patterns in Ds and Da domains of ADAMalysin family proteins affect the relative orientations of M and C domains. (A) The disulfide pattern in the D domain causes great conformational changes of two SVMPs. In the atragin structure, there are totally eight disulfide bonds in the D domain (numbers in black), of which C434–C457 and C453–C479 are enclosed in blue circles, whereas in the K-like structure, one new disulfide bond C434–C479 is formed and enclosed in red. The absent segment of K-like is colored in red in the atragin structure. (B) Sequence alignment of Ds and Da domains derived by ADAM/adamalysin/reprolysins family proteins and disintegrins. Above the black line, protein sequences with the same cysteine arrangement are aligned, whereas, below the line, protein sequences with significantly different disulfide bond pattern are aligned. The location of disintegrin-loop (XXCD) is labeled in yellow and the green number represents the disulfide pairs in the atragin structure. Bitistatin and human ADAM10 are highlighted by the shadow because their disulfide pairs are also different from conventional ones (e.g., atragin) despite of the same arrangement of csyteins with ADAMs. (C) Four protein structures with different disulfide bond patterns of D domains have distinct orientations of putative metalloprotease domains. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)