Figure 1.
The Cloverleaf Is an Important cis Active Element for Negative Strand RNA Synthesis
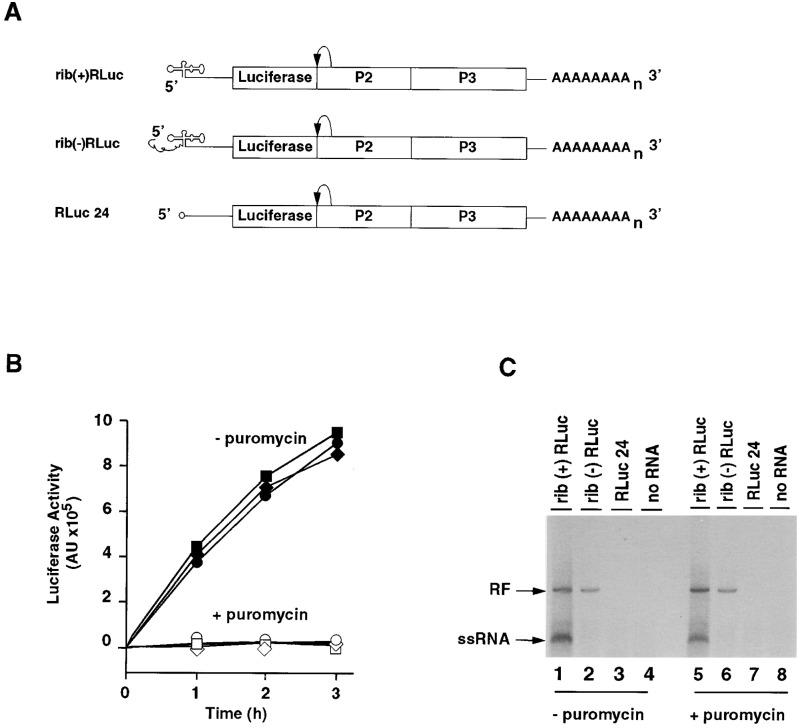
(A) Schematic representation of three replicons used, in which the structural protein coding region was replaced by the luciferase gene. Luciferase is released from the poliovirus polyprotein by the proteolytic activity of 2Apro. Replicon rib(+)RLuc RNA carries authentic 5′ ends; in contrast, the rib(−)RLuc replicon contains an extension of 50 nucleotides that prevents positive strand RNA synthesis in vitro (Herold and Andino, 2000). RLuc24 carries a deletion of the entire 5′ cloverleaf RNA. This construct contains a 5′-terminal cap structure (o-). All three constructs had a poly(A) tail of 80 adenylates.
(B) Translational activity of the replicon RNAs in vitro. HeLa extracts were programmed with the replicon RNAs (rib[+]RLuc, squares; rib[−]RLuc, diamonds; RLuc24, circles). Translation was efficient in the absence of puromycin (black symbols). However, luciferase activity was not observed in the presence of puromycin (negative control, white symbols).
(C) RNA synthesis in the absence and presence of puromycin. Translation/replication extracts were programmed with the replicon RNAs. After isolation of preinitiation complexes, the reactions were divided into two aliquots, and RNA synthesis reaction was performed either in the absence (lanes 1–4) or the presence (lanes 5–8) of 100 μg/ml puromycin. The products of the reaction were analyzed by native 1% agarose gel electrophoresis