Fig. 1.
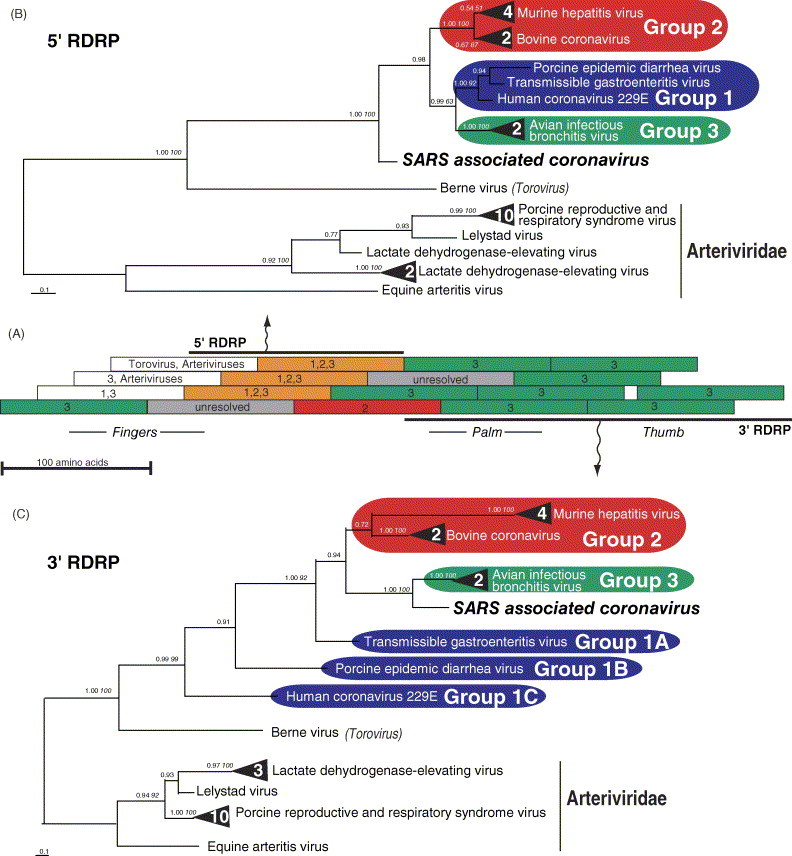
Recombinant nature of SARS-CoV RNA dependent RNA polymerase, as indicated by different sister relationships with other coronaviruses for different gene regions. (A) Schematic diagram showing Bayesian inference (BI) sliding window (100 amino acids long in 25 amino acid intervals) analyses used to assess recombination breakpoints within RDRP. The sister relationship of SARS-CoV RDRP with other coronavirus groups (1, 2 and/or 3) for each fragment is indicated by color code and numbers. BI phylogenies are shown for the entire 5′ (B) and 3′ (C) regions of RDRP. Numbers by each node are posterior probabilities, and when applicable, are followed by neighbor joining bootstrap percentages in italics. Multiple terminal nodes from a single virus species are represented by a black triangle, with the number of terminals indicated in white numerals. GenInfo identifiers for the proteins used in this analysis: 482297, 564004, 7769353, 93916, 233625, 14917044, 6625761, 13752450, 10242469, 94017, 12744851, 10179430, 25121660, 20271248, 11878197, 7650194, 17529672, 11878201, 25361011, 26008080, 12082740, 133455, 29293454, 14250963, 12240326, 10181074, 9635157, 29837504.
