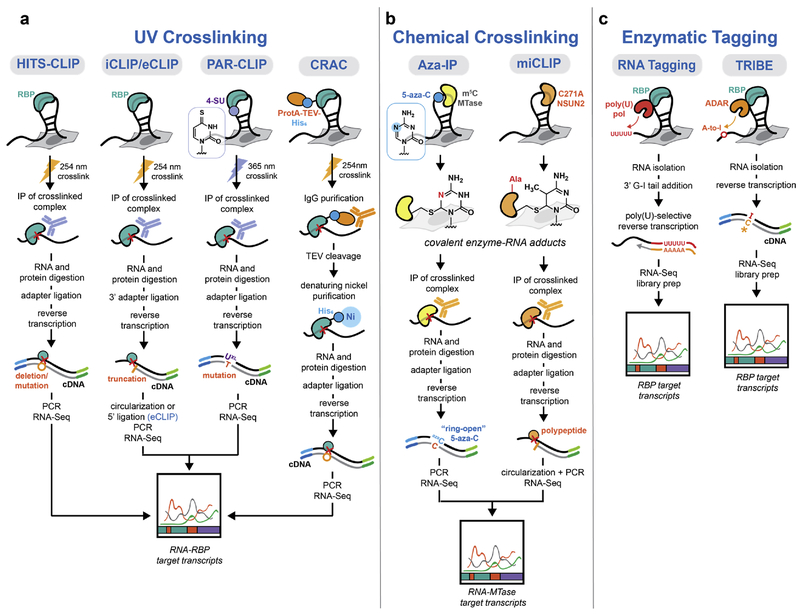
Figure 1.
Methods to identify RNA-protein binding sites. (a) Comparison of UV crosslinking-based CLIP methods. RBP footprints are identified by UV crosslinking, immunoprecipitation, and sequencing of covalently linked RNA. Variations on the prototypical HITS-CLIP protocol have been developed including modified photocrosslinking (PAR-CLIP), cDNA library generation (iCLIP/eCLIP), and immunoprecipitation (CRAC). (b) Chemical crosslinking approaches take advantage of the catalytic mechanism of RNA m5C methyltransferases, which involves formation of a covalent adduct with RNA substrates. In the Aza-IP strategy, metabolic incorporation of 5-azacytidine generates stalled covalent adducts at sites of modification. In miCLIP, a mutant RNA m5C methyltransferase lacking the ability to release itself from RNA substrates generates covalently bound complexes. (c) Non-crosslinking methods to map RBP substrates. In RNA Tagging, 3’ poly(U) tails are deposited on RBP-associated RNA by a poly(U) polymerase, PUP-2, fused to the RBP of interest. Following RNA isolation, target transcripts are enriched by reverse transcription with a primer specific for uridylated RNAs. TRIBE utilizes an RBP-ADAR construct to label associated RNA transcripts through adenosine to inosine (A:I) editing, which can then identified by RNA sequencing analysis (I pairs to predominantly to C).