Fig. 1.
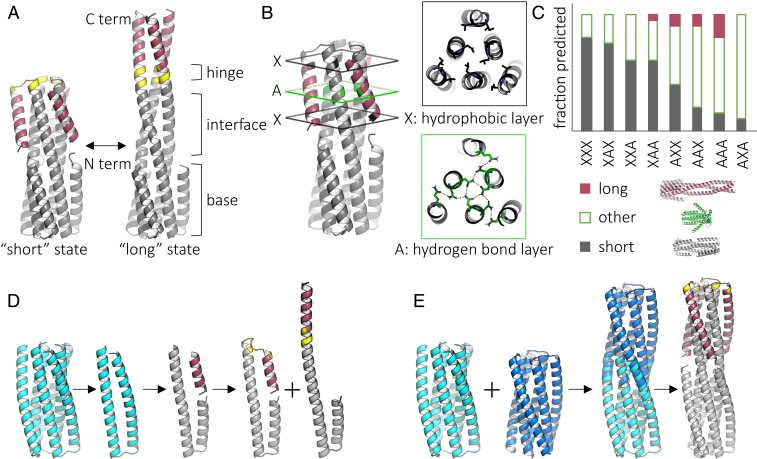
Design and in silico characterization of proteins with two distinct, well-defined ground states. (A) Design concept for protein backbones with a “short” and a “long” state. The backbone consists of a stable base, mobile flipping helices (red), tunable interface, and a flexible hinge (yellow). (B) Tunable interface between the inner helices and flipping helices. Each of three positions can be a hydrogen bond (green, “A”) or hydrophobic (black, “X”) layer. (C) Fraction of top 10 scored Rosetta folding predictions that resemble the short, long, or other structure for each permutation of possible interface configurations. (D) Initial design scheme. Starting with the previously characterized protein 2L3HC3_13 (PDB ID 5J0H) (cyan), a monomer is extracted, cut to produce a flipping helix, and reconnected with a hinge to produce the backbone of the short or long state. (E) Final design scheme. Previously characterized proteins 2L3HC3_13 (cyan) and 2L3HC3_23 (blue) are fused via their inner helices, outer helices are trimmed, and the hinge length is chosen such that the flipping helices pack against each other in the long state.