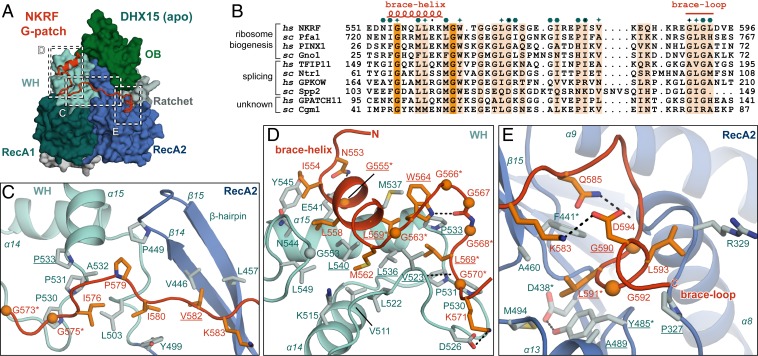
Fig. 2.
Conserved G-patch residues are crucial for forming hydrophobic interactions. (A) Overview of the complex with three different areas of G-patch interaction with DHX15 indicated by dotted frames, which are displayed in detail in C–E. Position D corresponds to G-patch site 1, while position E indicates G-patch site 2, and C marks the intervening linker. (B) Sequence alignment of G-patch proteins from yeast and human that have been assigned as activators of DHX15/Prp43 or DHX16/Prp2. Respective hs and sc homologs are grouped, and the corresponding cellular process is indicated. Brace-helix and brace-loop positions as found in NKRF are labeled on top of the alignment. Dark and light orange highlight residues that are identical and more than 70% similar, respectively. Petrol hexagons mark amino acids involved in DHX15 interactions, while stars mark mutated residues. (C) Interface of the G-patch linker region crossing over to RecA2 via the β-hairpin. (D) Interactions of the N-terminal G-patch brace-helix (red/orange) on the WH domain (cyan/light blue). Names of residues mutated in this study are underlined, and residues mutated in previous studies are marked by asterisks. Hydrogen bonds are indicated by black dotted lines. (E) Binding of the C-terminal G-patch brace-loop to the RecA2 domain.