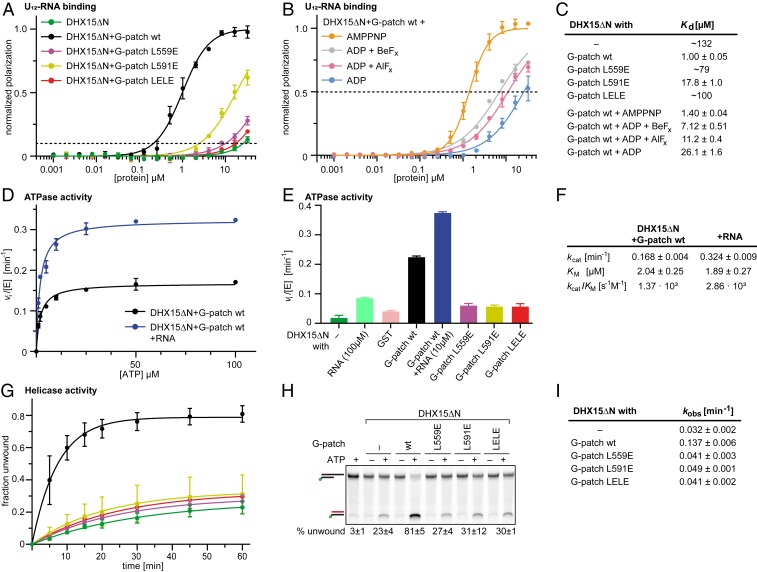
Fig. 5.
NKRF G-patch–mediated enhancement of DHX15 RNA affinity and ATPase activity depends on efficient domain tethering. (A) Fluorescence polarization experiments of FAM-U12 RNA with His10–DHX15ΔN in the absence or presence of GST–NKRF G-patch wt or mutants. The dashed line indicates 10% normalized polarization. Average values of triplicate measurements were plotted, and error bars correspond to SDs. (B) Fluorescence polarization experiments with DHX15ΔN and GST-NKRF G-patch wt in the presence of different ATP analogs or ADP (100 µM). The dashed line indicates 50% normalized polarization. Average values of triplicate measurements were plotted, and error bars correspond to SD. (C) RNA dissociation constants (Kd) with SEM determined by fitting curves in A and B by linear regression. For curves that do not reach 50% normalized polarization, the fitted constants were rounded to two digits and listed as approximate values. (D and F) Michaelis–Menten plot of ATPase activity of His10–DHX15ΔN in complex with GST–NKRF G-patch wt with (blue) or without (black) RNA. Measurements were carried out in triplicate, and error bars represent SD. Michaelis–Menten parameters and corresponding SEM were determined by linear regression. (E) Initial ATP hydrolysis rates at 2 mM ATP, normalized for enzyme concentrations for His10–DHX15ΔN with or without GST–NKRF G-patch wt or mutants, or GST as a control. Error bars indicate SD of triplicate measurements. (G and I) RNA duplex unwinding assays with His10–DHX15ΔN in the presence or absence of GST–NKRF G-patch wt or mutants using FAM-labeled dsRNA with a 3′ single-stranded overhang. Time courses were measured in triplicate, and error bars indicate SD. Rate constants and respective SEM were determined by linear regression. (H) Native PAGE of representative samples from the unwinding assay after 60-min reaction time detecting FAM-fluorescence signal. Samples without ATP or without protein were used as controls for RNase contamination. The top bands correspond to duplex RNA substrate, while the lower bands stem from unwound RNA that is quenched with a cDNA oligo.