-
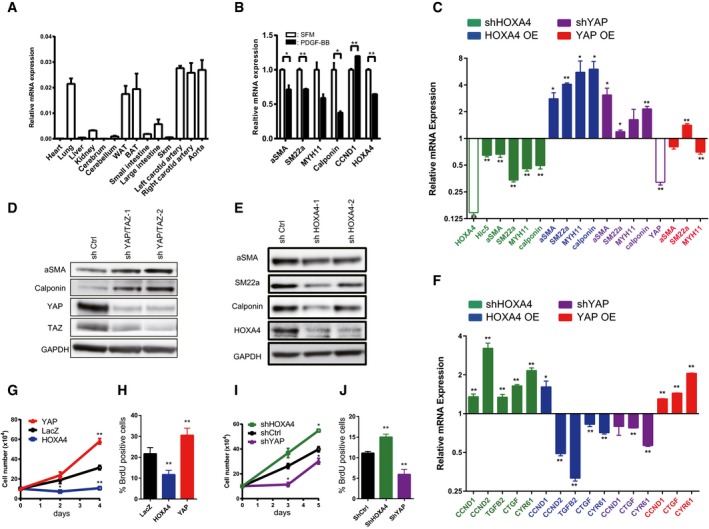
A
Expression of Hoxa4 in various mouse tissues using qRT–PCR. Expression of glyceraldehyde‐3‐phosphate dehydrogenase (Gapdh) mRNA was used as an internal control. WAT; white adipose tissue, BAT; brown adipose tissue, Skm; skeletal muscle.
-
B
Quantitative real‐time PCR analysis of differentiated VSMC marker genes in primary human VSMCs with PDGF‐BB stimulation for 24 h. *P < 0.05, **P < 0.01, by unpaired two‐tailed Student's t‐test.
-
C
Quantitative real‐time PCR analysis of smooth muscle‐specific genes in primary human VSMCs transduced with lentiviral vectors encoding overexpression or knockdown of HOXA4 and YAP. Cells infected with lentiviral vectors encoding LacZ or shCtrl were used as controls. *P < 0.05, **P < 0.01, by unpaired two‐tailed Student's t‐test.
-
D, E
Representative Western blotting analysis of total protein lysates of primary human VSMCs transduced with lentiviral vectors encoding knockdown of YAP/TAZ (D) or HOXA4 (E). Representative Western blotting analysis of total protein lysates of primary human VSMCs transduced with lentiviral vectors encoding knockdown of HOXA4.
-
F
Quantitative real‐time PCR analysis of YAP/TEAD target genes in primary human VSMCs transduced with lentiviral vectors encoding overexpression or knockdown of HOXA4 and YAP. The cells infected with lentiviral vectors encoding LacZ or shCtrl were used as controls. *P < 0.05, **P < 0.01, by unpaired two‐tailed Student's t‐test.
-
G
Proliferation assay in VSMCs transduced with the indicated genes. Virus‐transfected human VSMCs were plated at equal density in growth medium, and then, cells were collected and counted at each time point as indicated. **P < 0.01 versus LacZ, by ANOVA with Sidak's correction.
-
H
BrdU incorporation assays in VSMCs transduced with the indicated genes. The percentage of BrdU‐positive cells in each group is shown. Nine to twelve images were randomly acquired and analyzed. **P < 0.01 versus LacZ, by ANOVA with Sidak's correction.
-
I
Proliferation assay in VSMCs transduced with the indicated genes. Virus‐transfected human VSMCs were plated at equal density in growth medium, and then, cells were collected and counted at each time point as indicated. *P < 0.05 versus shCtrl, by ANOVA with Sidak's correction.
-
J
BrdU incorporation assays in VSMCs transduced with the indicated genes. The percentage of BrdU‐positive cells in each group is shown. Nine to twelve images were randomly acquired and analyzed. **P < 0.01 versus shCtrl, by ANOVA with Sidak's correction.
Data information: Graphs are presented as means ± SEM; three biological repeats, except for (H) and (J).