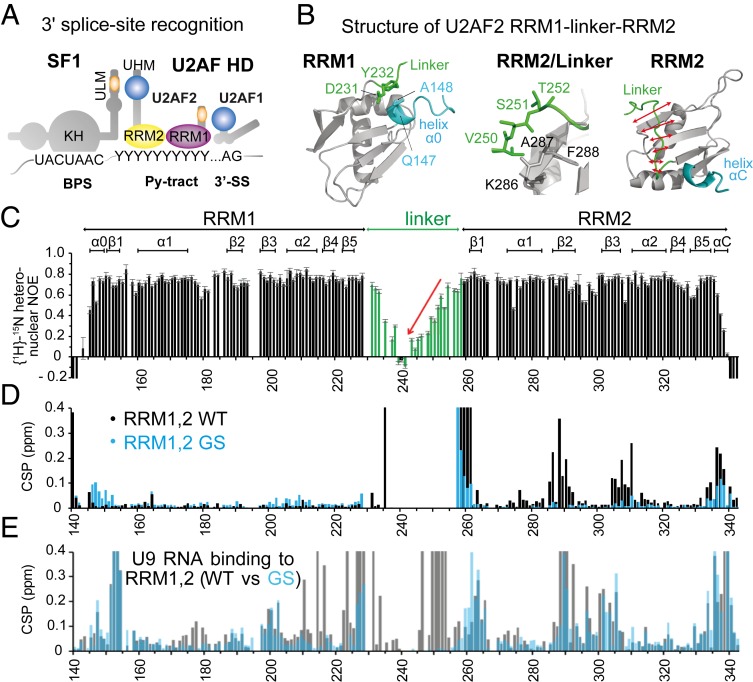
Fig. 1.
Overview of 3′ splice site recognition and structural features of U2AF2 RRM1,2. (A) Overview of cis-regulatory elements (BPS, Py-tract, AG dinucleotide) and splicing factors (SF1, U2AF2, U2AF1) that cooperate in 3′ splice site recognition in human introns. Asterisks denote phosphorylation sites. (B) NMR structures of U2AF2 RRM1 and RRM2 obtained for the redefined U2AF2 RRM1,2 protein (residues 140 to 342). RRM1 (Left) is preceded by a short helical turn (α0), which is in proximity to the N-terminal residues of the RRM1,2 linker, also showing reduced flexibility. RRM2 (Right) shows that the C-terminal region of the linker packs against the β-sheet surface that is also involved in RNA binding. RRM2 has a short C-terminal helical turn (helix αC, blue). (C) NMR {1H}-15N heteronuclear relaxation data for U2AF2 RRM1,2 show that the central linker is highly flexible, but exhibits increased rigidity as it approaches the N-terminal end of the RRM2 fold. (D) Chemical shift differences comparing wildtype (WT; black) and the linker GGS mutation (GS; cyan) of U2AF2 RRM1,2 vs. the individual RRM1 and RRM2 domains. (E) Chemical shift perturbation for WT (black) and GS (cyan) U2AF2 RRM1,2 upon addition of U9 RNA.