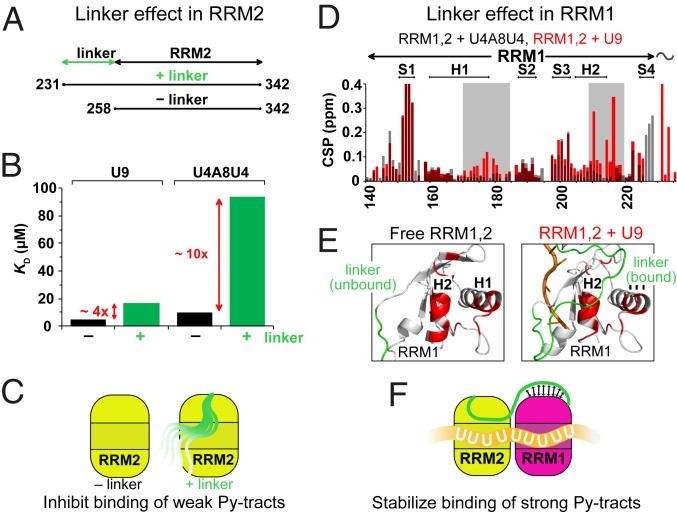
Fig. 3.
Molecular features that enable proofreading of U2AF2 RRM1,2 against weak Py-tracts. (A) Constructs used (RRM2 and linker–RRM2) to study the role of the linker. (B) Dissociation constants KD for the binding of RRM2 and linker–RRM2 to strong (U9) and weak (U4A8U4) Py-tract RNAs determined by ITC. (C) Role of the C-terminal region of the RRM1,2 linker: the C-terminal region of the linker competes with binding of weak Py-tracts. (D) NMR chemical shift perturbations (CSPs) in the RRM1 region upon binding of RRM1,2-WT to strong (U9) and weak (U4A8U4) Py-tract RNAs (cf. SI Appendix, Fig. S5). Significantly stronger CSPs are observed for residues (gray box) that contact the RNA upon binding to strong Py-tract RNA. (E) The N-terminal region of the RRM1,2 linker is flexible in the unbound protein, but weakly stabilizes RNA interactions when bound to a strong Py-tract RNA. The distinct CSPs upon binding to the strong Py-tract RNA shown in D are highlighted in red on a cartoon presentation of RRM1. (F) Role of the N-terminal region of the RRM1,2 linker: the N-terminal region of the linker may stabilize binding of strong (U-rich) Py-tract RNAs.