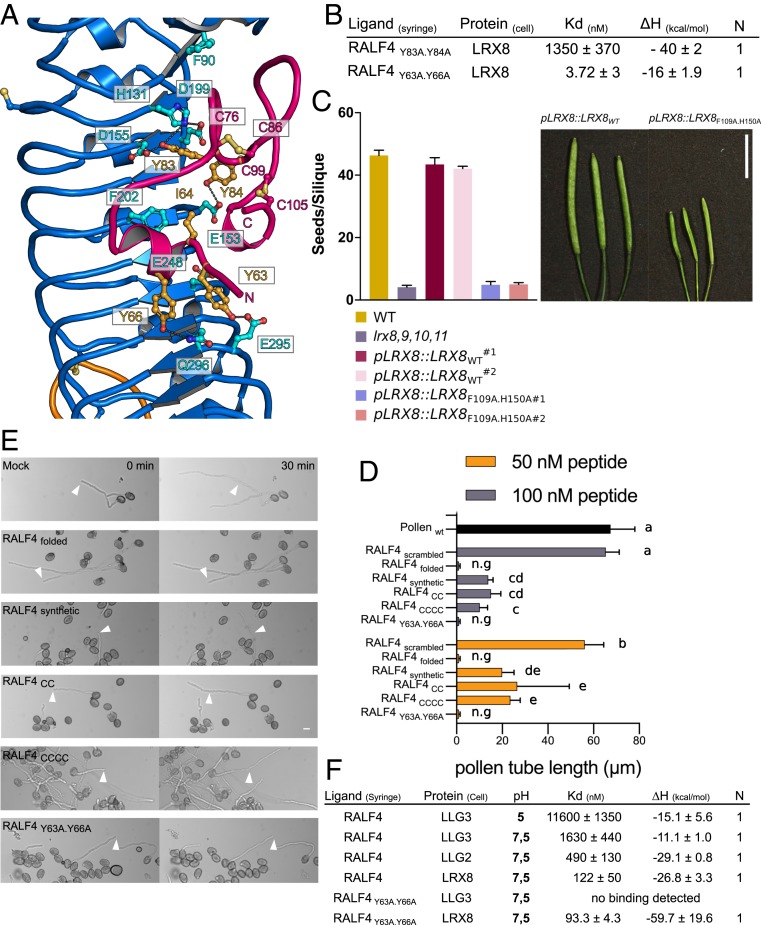
Fig. 3.
LRX8 bioactivity depends on the interaction with folded RALFs. (A) Close-up view of the LRX2–RALF4 binding pocket. A large surface of the RALF4 peptide (in pink) directly interacts with the LRR core (in blue) of LRX2. Polar contacts of RALF4 with LRX2 are shown as dotted lines (in gray). The two disulfide bridges responsible for the folding of the peptide, C76–C86 and C99–C105, are depicted as sticks in light orange. (B) ITC table summaries of LRX8 vs. RALF4Y83A.Y84A and RALF4Y63A.Y66A mutants. Table summaries show the dissociation constant (Kd), binding stoichiometries (N), and thermodynamic parameters. The values indicated in the table are the mean ± SD of two or three independent experiments. (C) Complementation assay of lrx8,9,10,11 with point mutated LRX8F109A.H150A driven by its endogenous promoter. (C, Left) Seeds per silique of WT, lrx8,9,10,11, pLRX8::LRX8WT lines 1 and 2, and pLRX8::LRX8F109A.H150A in the lrx8,9,10,11 quadruple mutant background, for two independent lines. Data are means ± SEM of 10 siliques. For LRX8 mutant transgenic lines, 16 independent lines were analyzed. (C, Right) Silique images of the corresponding lines. (Scale bar, 10 mm.) (D) Growth rate of wild-type pollen tubes after the addition of RALF4scrambled, RALF4folded, RALF4synthetic, RALF4CC, RALF4CCCC, and RALF4Y63A.Y66A peptides at different concentrations after 30 min. Data were analyzed by one-way ANOVA followed by Tukey’s test as post hoc, considering P ≤ 0.05 as significantly different; data shown are mean ± SEM of three biological replicates, n = 28 each. Same letters represent samples that are not different between each other; n.g, no growth after peptide addition. (E) Effect of RALF4 peptides on wild-type pollen tubes growing in semisolid medium at two time points after adding them to a concentration of 50 nM. Arrowheads indicate the position of the tip of selected pollen tubes at time point 0 min. (Scale bar, 10 µm.) (F) Binding matrix of RALF4folded and RALF4Y63A.Y66A peptides vs. LRX8 and LLG2/3 membrane proteins in acidic and alkaline conditions. ITC table summary with dissociation constants (Kd), binding stoichiometries (N), and thermodynamic parameters (ΔH).