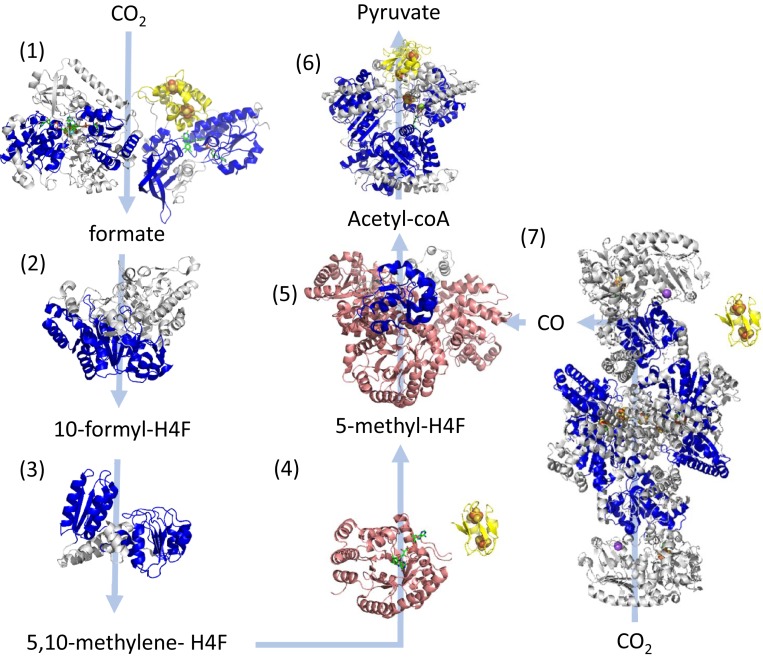
Fig. 3.
Structurally determined oxidoreductases in the Wood–Ljundhal carbon-fixation pathway from CO2 to pyruvate performed by early acetogens (26). Modules within these proteins are colored as follows: blue, Rm folds; yellow, Fd folds; and pink, β/α barrels. Gray domains designate other modules or unassigned structural motifs. Where possible, the locations of cofactors are included in the structure seen as spheres for metals or as green sticks for organic cofactors. Light blue arrows indicate substrate-product transformations catalyzed by each enzyme. Numbers designate each of the reactions and the structures of enzymes catalyzing these reactions are the following: reaction 1—NADPH formate dehydrogenase (PDB ID codes 1FDO.A and 4YRY.B); reaction 2—10-formyl-H4 folate synthetase (PDB ID code 4IOJ); reaction 3—bifunctional protein FolD including methylene tetrahydrofolate dehydrogenase and methenyl tetrahydrofolate cyclohydrolase (PDB ID 5A5O); reaction 4—5,10-methylene-H4 folate reductase (PDB ID code 3APT); reaction 5—corrinoid iron-sulfur protein (CFeSP) (PDB ID code 4DJD); reaction 6—pyruvate synthase (PDB ID code 6CIN); and reaction 7—CO dehydrogenase/acetyl-CoA synthase (PDB ID code 3I01). The enzymes shown in groups 4 and 7 utilize the electrons from a soluble ferredoxin, which is not in the experimental structure and is illustrated using the bacterial ferredoxin structure PDB ID code 1FDN.