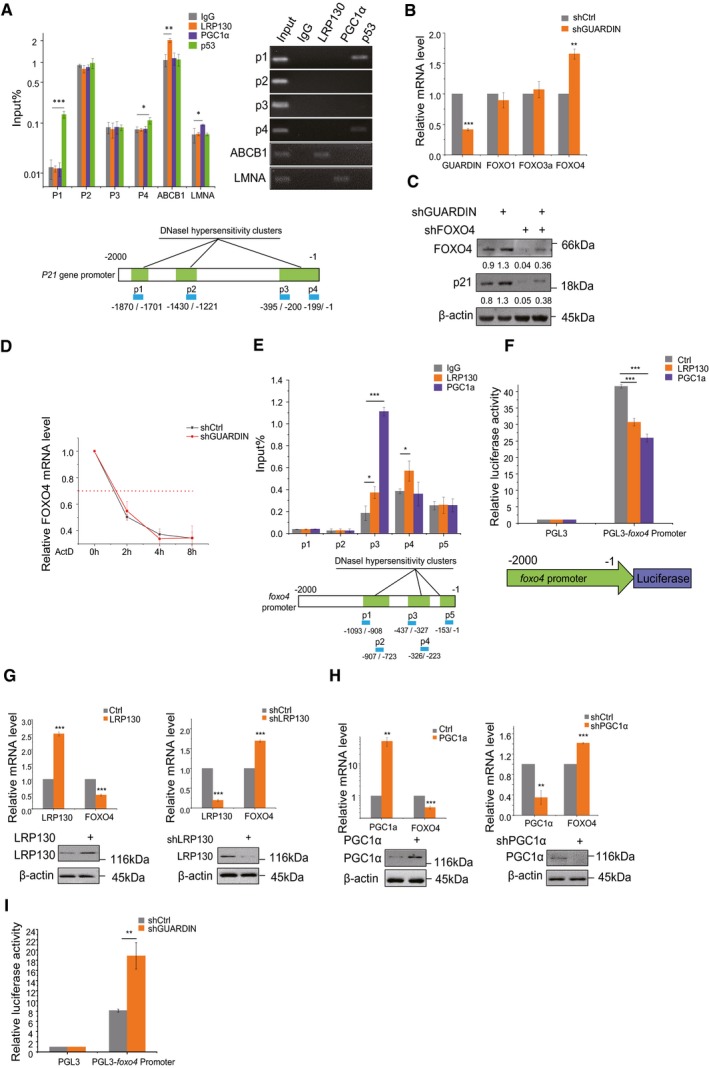
ChIP assays detecting binding of LRP130/PGC1α to the p21 promoter using qPCR and RT–PCR (top left and right, respectively). IgG and p53 served as a negative and positive controls, respectively. Schematic illustrations of the putative LRP130/PGC1α binding sites within DNase I hypersensitive regions of the p21 promoter (bottom).
qPCR assays for GUARDIN, FOXO1, FOXO3a, and FOXO4 in A549 cells after 24‐h transduction with shCtrl or shGUARDIN.
Western blotting assays for FOXO4 and p21 protein expression in A549 cells after 48‐h transduction with shCtrl or shGUARDIN alone or in combination with shFOXO4. β‐actin served as loading control.
Half‐life times of FOXO4 mRNA in A549 cells with shCtrl or shGUARDIN measured by qPCR after treating cells with 5 μg/ml of actinomycin (ActD) for the indicated times.
ChIP assays detecting binding of LRP130/PGC1α to putative binding sites in the FOXO4 promoter using qPCR (top). Data were normalized to the IgG negative control. Schematic illustration of the LRP130/PGC1α binding sites within DNase I hypersensitive regions of the FOXO4 promoter (bottom).
Luciferase assays conducted in A549 cells transfected with LRP130 or PGC1α using the pGL3 (negative control) or pGL3‐FOXO4 promoter reporter plasmids.
qPCR assays for LRP130 and FOXO4 mRNA levels (upper) and Western blotting analysis for LRP130 protein level (lower) in A549 cells after 48‐h transduction with LRP130 overexpression or knockdown.
qPCR assays for PGC1α and FOXO4 mRNA level (upper) and Western blotting analysis for PGC1α protein level (lower) in A549 cells after 48‐h transduction with PGC1α overexpression or knockdown.
Luciferase activity assays in A549 cells transduced with shCtrl or shGUARDIN using the pGL3 (negative control) or pGL3‐FOXO4 promoter reporter plasmids.
= 3 biological replicates). (A, B, E–I) two‐tailed paired Student's
< 0.001).