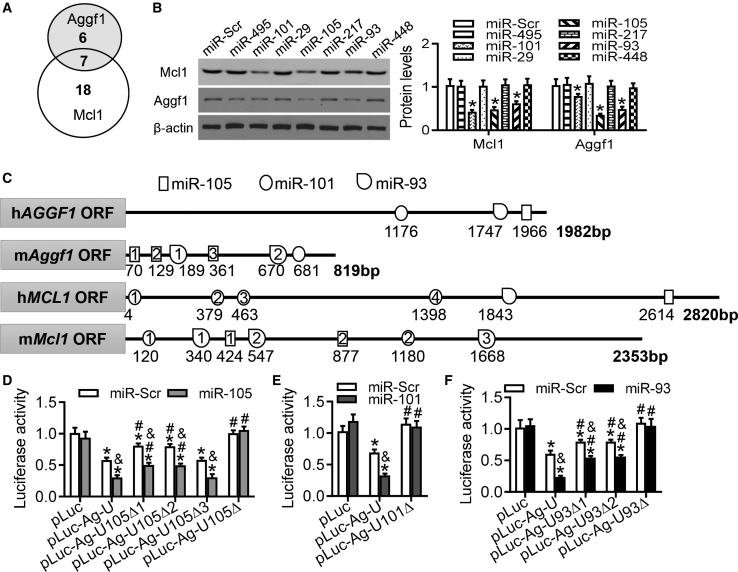
Figure 5.
Characterization of miRNAs in the 3′ UTRs of Aggf1 and Mcl1 mRNAs
(A) Summarization of predicted MREs in the 3′ UTRs of Aggf1 and Mcl1. (B) Western blot analysis for Aggf1 and Mcl1 expression was performed after overexpression of different miRNA candidates in purified mouse neocardiomyocytes. *p < 0.05 versus miR-Scr. (C) Bioinformatic analysis of the Aggf1 and Mcl1 mRNA sequences identified binding sites for miR-105, miR-101, and miR-93 at the 3′ UTRs. (D) HEK293 cells were transfected with different luciferase plasmids and miR-Scr or miR-105. The luciferase activities were measured. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr. (E) HEK293 cells were transfected with different luciferase plasmids and miR-Scr or miR-101. The luciferase activities were measured. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr. (F) The luciferase activities were measured in HEK293 cells transfected with different luciferase plasmids and miR-Scr or miR-93. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr.