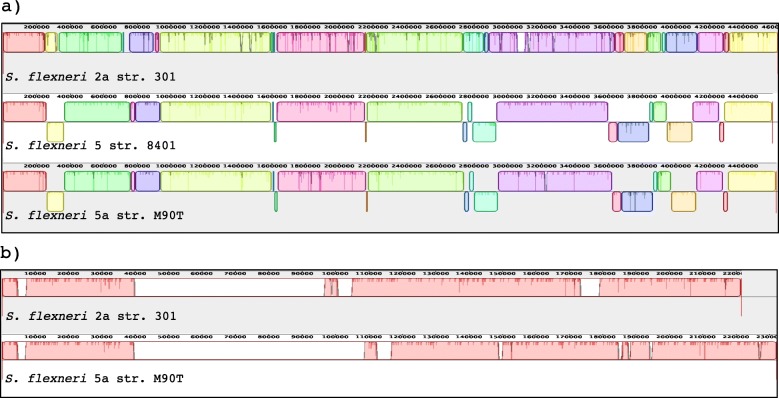
Fig. 2.
Comparative genomic map of sequenced S. flexneri strains. a Chromosome comparison of S. flexneri 2a 301 (NC_004337), S. flexneri 5 8401 (NC_008258) and S. flexneri 5a M90T (NC_CP037923), b Virulence plasmid comparison of S. flexneri 2a 301(NC_004851) and S. flexneri 5a M90T (NZ_CP037924). Genome-wide alignment was performed with Mauve [56] progressive alignments to determine conserved sequence regions. This alignment resulted in many large synteny locally collinear blocks (LCBs). Each syntenical placement of the homologous region of the genome is represented as unique colored block, whilst divergent regions are seen as an empty block or line. Indentations within boxes highlight small mutations. Blocks above and below the center line depict the orientation of the genomic region compared to S. flexneri 2a strain 301