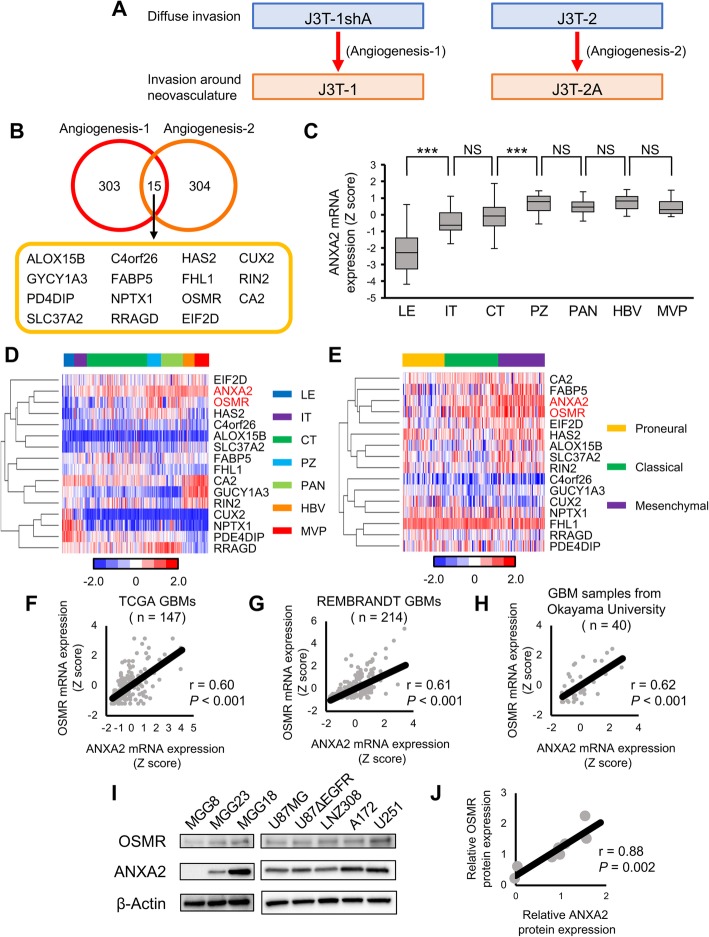
Fig. 1.
Identification of candidate ANXA2-regulated genes that promote angiogenesis and cell invasion. a Microarray analysis was performed to identify ANXA2-regulated genes that promote the angiogenesis–invasion phenotype. See text for the definitions of Angiogenesis-1 and Angiogenesis-2 genes. b Venn diagram of Angiogenesis-1, Angiogenesis-2, and shared genes identified from the analysis in (a). c Anatomical expression pattern of ANXA2 mRNA in the brain from the Ivy Glioblastoma Atlas Project dataset (n = 270). LE, leading edge; IT, infiltrating tumor; CT, cellular tumor; PZ, perinecrotic zone; PAN, pseudopalisading cells around necrosis; HBV, hyperplastic blood vessels; MVP, microvascular proliferation. Data are shown as the mean ± SEM. ***P < 0.001, NS not significant by one-way ANOVA with Bonferroni’s post hoc test. d Hierarchical clustering of genes related to angiogenesis and invasion ordered by anatomical location (Ivy GAP dataset, n = 270). e Hierarchical clustering of genes related to angiogenesis and invasion ordered by GBM molecular subtypes GBM (TCGA dataset, n = 143). f–h Pearson’s correlation tests of ANXA2 and OSMR mRNA expression in GBM datasets from TCGA (F, n = 147) and REMBRANDT (g, n = 214) and from GBM samples from Okayama University (h, n = 40). i Western blot analysis of ANXA2, OSMR, and β-actin in established human GBM cell lines and patient-derived GBM cells. j Pearson’s correlation test of ANXA2 and OSMR protein expression from the data shown in (i)