Figure 4.
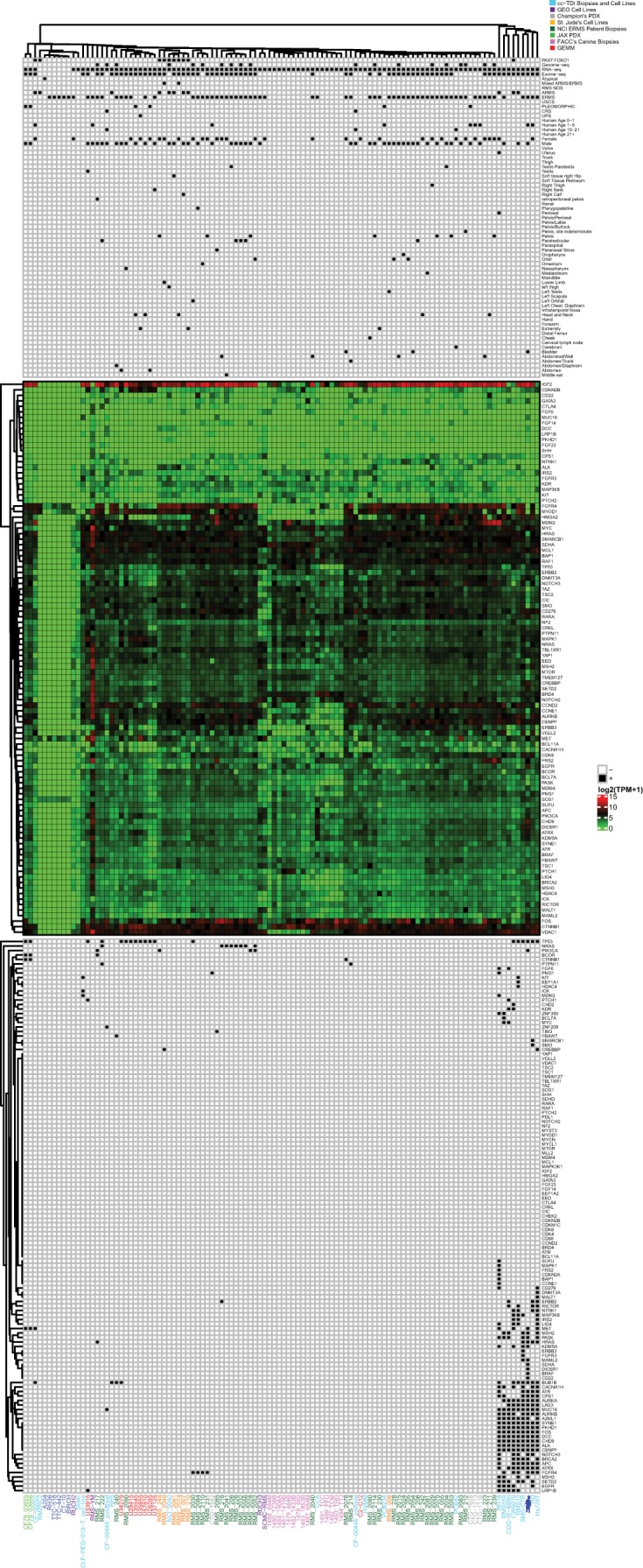
Index case endotype unsupervised clustering of embryonal rhabdomyosarcoma (ERMS) and non-rhabdomyosarcoma soft-tissue sarcoma (NRSTS) samples with alveolar rhabdomyosarcoma (ARMS) samples harboring the PAX7:FOXO1 fusion gene as a control determined several endotypes. Clustering was performed using DNA and RNA sequencing from tumor samples from our IRB-approved CuReFAST initiative and cell lines (light blue), cell lines from Gene Expression Omnibus (GEO) (purple), PDX mouse models from Champions Oncology (light gray), cell lines from St. Jude Children's Research Hospital (blue), ARM patient's biopsies from National Cancer Institute (orange), ERMS patient's biopsies from NCI (dark green), PDX mouse models from the Jackson Laboratory (light green), canine samples from Flint Animal Cancer Center (pink), genetically engineered mouse models (red), and the patient's tumor sample (dark blue avatar). The legend below marks samples with known sex, age, primary tumor site, somatic mutations, and diagnosis. Gene expression (log2 (TPM +1)) is shown below in a heatmap on a scale of 15 (red) to 0 (green). Samples without matched RNA-sequencing data were given a value 0 for all genes. Below the gene expression heatmap, samples with somatic mutations in our genes of interest are indicated by a black box. Unsupervised clustering was performed separately on the expression and somatic data within the legend (vertical dendrogram). Because we suspected a germline predisposition/syndrome for this patient, we kept germline mutations in the dendrogram but did not do the same for the other tumors as they were presumably somatic, noninherited cases.
