Abstract
The helicase-like domain of the Bamboo mosaic virus replicase catalyzes the release of 5′-γ-phosphate from both ATP and 5′-triphosphated RNA by an identical set of catalytic residues with a presumably larger binding pocket for RNA. In this study, the peptidyl regions involved in RNA binding were mapped by reversible formaldehyde crosslinking and mass spectrometry. Eleven residues within these regions were examined by mutational analysis. H636A, Y704A, and K706A greatly diminished the enzymatic activities and were unable to support the viral replication in Nicotiana benthamiana protoplasts. K843A decreased activity toward the RNA substrate to 17% of WT, and ∼ 20% replication efficiency was retained in protoplasts. R597A and K610A retained ∼ 50 and ∼ 90% of the enzymatic activities, respectively. However, replication in protoplasts of these mutants was extremely limited. Proteins with the mutations K603A, R628A, R645A, H794A, and R799A were present at levels 30–69% of WT in protoplasts. However, the fates of these mutations in plants were different. Viral cell-to-cell movement was limited by the K603A and R628A mutations, while systemic movement was restricted by R645A and H794A. The implications of the helicase-like domain in the viral replication and movement are discussed.
Keywords: RNA-binding, 5′-Triphosphatase, Helicase, Bamboo mosaic virus, Reversible formaldehyde crosslinking
Helicases are traditionally defined as a class of enzymes that catalyze the separation of duplex nucleic acids into single strands in an ATP-dependent reaction. They actively participate in a variety of biological functions including DNA replication, DNA repair, recombination, transcription, translation, and many other nucleic acid-related processes (Jankowsky and Fairman, 2007). Based on the number and amino acid sequence of the NTP-binding helicase motifs, the enzymes have been divided into three superfamilies and two smaller families (Gorbalenya and Koonin, 1993). The helicases (or helicase-like domains) encoded by RNA viruses can be placed into one of the three superfamilies, regardless of whether they catalyze a bona fide duplex unwinding event (Kadaré and Haenni, 1997). The activity of accelerating NTP hydrolysis (NTPase) is the common feature of the viral helicases. In addition, many of them, such as those from reovirus (Bisaillon and Lemay, 1997), alphavirus (Vasiljeva et al., 2000), dengue virus (Bartelma and Padmanabhan, 2002), coronavirus (Ivanov et al., 2004), and potexvirus (Li et al., 2001), exhibit RNA 5′-triphosphatase (RTPase) activity, which is responsible for the removal of 5′-γ-phosphate from the nascent viral mRNA. Viral helicases with RTPase activity are thus considered essential for the formation of 5′-cap structure, important for translational initiation and mRNA stability.
Viral RNA helicases may be crucial for viral infectivity via mechanisms other than NTP hydrolysis and duplex unwinding. The helicase-like domain of Brome mosaic virus (BMV), a member of the RNA helicase superfamily 1 (SF1), represents an extensively studied example. BMV 1a protein, which contains AdoMet-dependent guanylyltransferase and helicase-like domains, selectively represses the translation of BMV RNA1 and RNA2 (Yi et al., 2007). Mutation of 1a at either the conserved R136 in the guanylyltransferase domain or at the conserved K691 in the helicase-like domain reduced the repression. 1a protein also recruits BMV 2a polymerase, RNA2 and RNA3 to form spherules on endoplasmic reticulum membranes, in which virus replication occurs (Chen and Ahlquist, 2000, Chen et al., 2001, Wang et al., 2005). Many of these functions depend on the presence of a B box, which is located in the 5′UTR of RNA2 and the intercistronic region of RNA3, implying that a probable binding between 1a and the specific RNA element is critical for BMV reproduction.
Bamboo mosaic virus (BaMV), a member of the potexvirus genus that belongs to the alphavirus-like superfamily, contains a ∼ 6.4-kb genome with a 5′-cap and a 3′-poly(A) tail (Lin et al., 1994). ORF1 of the viral genome encodes a 155-kDa polypeptide consisting of, from N to C terminus, an AdoMet-dependent guanylyltransferase, a helicase-like domain, and an RNA-dependent RNA polymerase. The BaMV helicase-like domain, also classified as SF1, exhibits ATPase and RTPase activities (Li et al., 2001). Inactivation of these two activities in response to mutations at the consensus residues within helicase motifs I, II, and VI and the competitive inhibition by an ATP analogue of RTPase activity suggest that the γ-phosphate of the two substrates is cleaved off by an identical set of catalytic residues (Han et al., 2007). However, it is reasonable to hypothesize that more residues are involved in RNA binding due to the much larger size of RNA than ATP. The question as to whether the BaMV helicase-like domain is able to unwind duplex RNA remains to be answered.
Inspired by the pivot roles of BMV 1a protein in assembling the replication complex and regulating the translation of viral proteins, we would like to know whether the BaMV helicase-like domain plays roles in various steps of viral replication via its RNA-binding ability. To elucidate how the helicase-like domain binds RNA, a reversible crosslinking technique using formaldehyde and mass spectrometry was used to map the RNA-binding regions in this study. Several peptidyl regions were identified and eleven selected residues within these regions were mutagenized to analyze their roles in RNA binding, phosphohydrolase activity, and in vivo viral replication and infectivity.
Results
Identification of potential RNA-binding residues
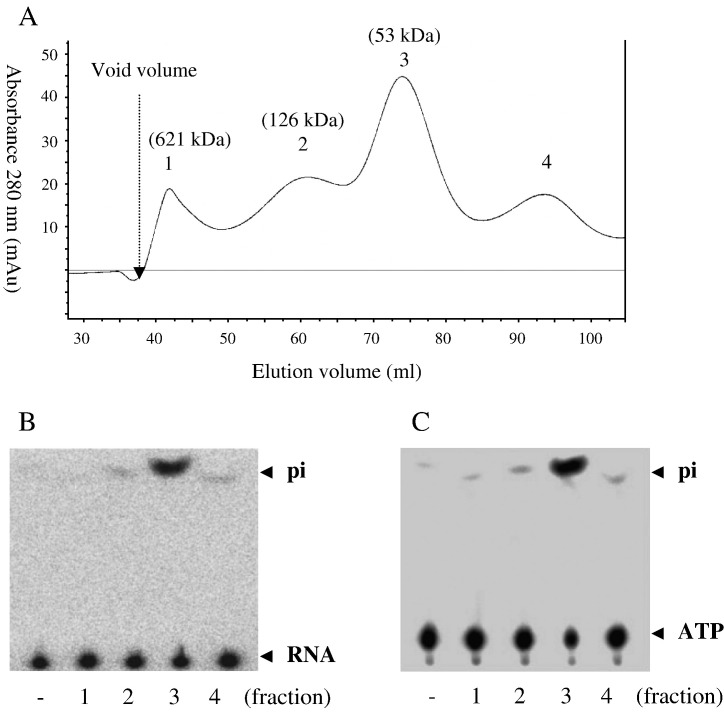
The BaMV helicase-like domain, generated as a fusion protein with a thioredoxin/hexahistidine/S-tag fused at the N-terminus, has been used to characterize associated enzymatic activities (Li et al., 2001, Han et al., 2007). Purification of the recombinant domain involved protein denaturation and refolding. To examine the oligomerization state of the refolded protein, a gel filtration step was included in the present study. According to the estimated molecular sizes, most refolded protein was in a monomeric form, while some associated with each other and formed dimers or probably even dodecamers (Fig. 1A). Activity assays indicated that the monomer has optimal RTPase and ATPase activities (Figs. 1B and C, respectively). Activity for unwinding duplex RNA was also surveyed for each of the protein forms since many viral helicases have been shown to function as hexa- or oligomers (Goregaoker and Culver, 2003, Levin and Patel, 1999, Uhlmann-Schiffler et al., 2002). However, no RNA unwinding activity was observed for any protein forms (data not shown).
Fig. 1.
Oligomerization states and activities of the BaMV helicase-like domain. (A) Gel filtration elution profile. The molecular mass of the refolded protein in each collected fraction (peaks 1–3) was estimated as described in Materials and methods. (B) RTPase. The protein in each collected fraction (30 ng each) was incubated with [γ-32p]RNA (0.4 μg) in a final 3 μl solution at 20 °C for 1 min. “−” indicates a control without addition of enzyme. The reaction products were resolved on TLC plates and visualized by autoradiography. (C) ATPase. The conditions were as in part B except that 0.2 mM ATP, containing 1 μCi [γ-32p]ATP, was used as the substrate.
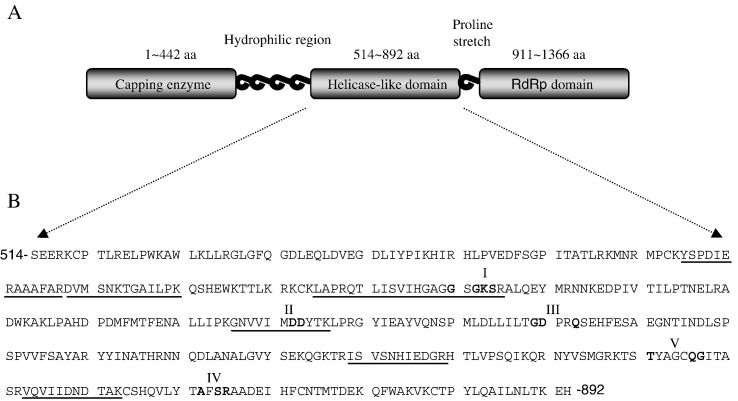
Reversible formaldehyde crosslinking is frequently utilized in protocols for assaying protein–DNA interactions in vivo, e.g. chromatin immunoprecipitation (Perez-Romero and Imperiale, 2007). It has also been adopted in the mapping of amino acid residues involved in the direct binding of proteins to nucleic acids (Kim et al., 2005). This technique relies on the formation of methylene bridges between the side chains of Lys, Arg, His, Cys, aromatic amino acids and bases (A, G, and C) of nucleic acids (Bernard et al., 2004). These crosslinks can be reversed via rehydration of the methylene bridges and release of formaldehyde by heating the sample; therefore, the binding partners can be characterized (Orlando et al., 1997). To further understand the structure–function relationship of the BaMV helicase-like domain, particularly with respect to its RNA-binding function, the protein in its monomeric form was incubated with a 3′-biotinylated 15-nt RNA in the presence of formaldehyde as described in Materials and methods. After digestion with trypsin, the peptides covalently linked to biotinylated RNA were affinity purified with avidin resin. After extensive washes, the crosslinked adduct was heated at 70 °C for 60 min to release the conjugated peptides, which were then subjected to sequence analysis by LC-MS/MS. To enhance the reliability of the data, the above experiment was repeated four times. Peptides repeatedly found to bind biotinylated RNA were selected and listed in Table 1 . They are distributed over the helicase-like domain, including two regions encompassing motifs I and II, respectively (Fig. 2 ). No peptide fragments deriving from the BaMV helicase-like domain were obtained in the parallel control reaction, in which the addition of formaldehyde was omitted. Several positively charged residues and a tyrosine within the selected peptidyl regions were then substituted with alanine for functional analysis based on their potential to interact with the RNA substrate. Eleven mutant proteins were expressed and purified. All had similar gel filtration profiles compared to WT.
Table 1.
Identification of peptidyl fragments bound to biotinylated RNA by LC-MS/MSa.
| Peptides (aa range) | Observedb | Mr (expt)b | Mr (calc)b | Deltab | Missb | Sequencec |
|---|---|---|---|---|---|---|
| 585–597 | 733.9446 | 1465.8746 | 1465.7313 | 0.1433 | 1 | YSPDIERAAAFAR |
| 598–610 | 687.4472 | 1372.8798 | 1372.7384 | 0.1415 | 1 | DVMSNKTGAILPK |
| 625–645 | 527.1123 | 2104.4200 | 2104.1864 | 0.2336 | 2 | LAPRQTLISVIHGAGGSGKSR |
| 696–706 | 627.8377 | 1253.6609 | 1253.5962 | 0.0647 | 0 | GNVVIMDDYTK |
| 789–799 | 409.5629 | 1225.6669 | 1225.6051 | 0.0618 | 0 | ISVSNHIEDGR |
| 833–843 | 608.3674 | 1214.7203 | 1214.6507 | 0.0697 | 0 | VQVIIDNDTAK |
The experiments were repeated four times and the peptides that appeared more than twice are listed. The amino acid sequences in the table were obtained from the second mass spectrometry.
The Miss represents the missed cleavage sites by trypsin on the identified peptides. The observed, Mr (expt), Mr (calc) and delta represent experimental m/z value, molecular mass transformed from experimental m/z, calculated molecular mass of the matched peptide, and difference between the experimental and calculated mass, respectively.
The residues selected for mutational analysis in this study are underlined. The consensus residues of motifs I (GSG) and II (DD) are shown in bold.
Fig. 2.
The peptidyl regions mapped to bind biotinylated RNA by formaldehyde crosslinking and LC-MS/MS analysis. (A) Domain organization of BaMV replicase. The central helicase-like domain, fused with a thioredoxin/hexahistidine/S-tag, was used to link the biotinylated RNA. (B) Amino acid sequence of the helicase-like domain. The underlined sequences indicate the mapped peptidyl segments. Consensus residues within each helicase motif are shown in bold.
Mutational effects on RNA binding
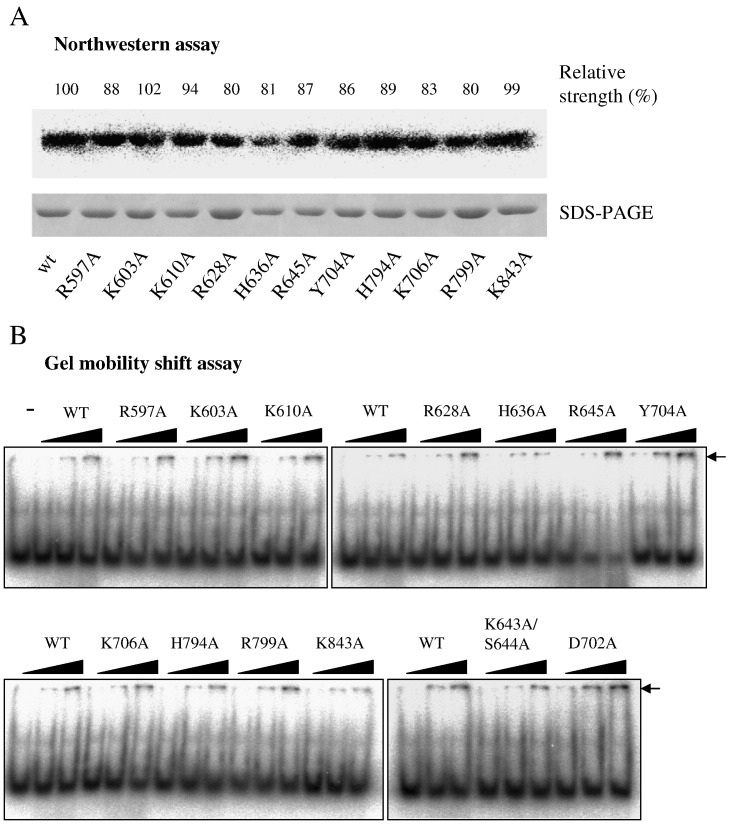
The effects on RNA-binding ability of each mutation were first examined by Northwestern assay (Fig. 3 ). After normalization to the amount of protein loaded, the mutant domains exhibited slightly decreased abilities to bind a 5′-terminal 178-nt genomic transcript compared to WT (Fig. 3A). It should be noted that the thioredoxin/hexahistidine/S-tag did not bind the RNA probe (data not shown). The binding ability was also assayed based on the mobility shift of a 50-nt RNA probe on native PAGE (Fig. 3B). Adding more amounts of the helicase-like domain in the reaction mixture would restrain more RNA molecules from migration. Once again, only minor effects on RNA binding were observed in response to the mutations. These results could be due to RNA-binding forces being contributed by every individual interaction, thus any single mutation would not significantly affect the overall binding strength. Alternatively, it is a possibility that none of the selected residues actually plays a role in RNA binding, however, this is an unlikely case. Previous studies showed that mutations at the consensus residues of motifs I and II abolished ATPase and RTPase activities (Han et al., 2007). The ability of the corresponding mutant proteins, K643A/S644A and D702A respectively, to bind RNA was also examined by gel mobility shift assay. Mutations at motif I slightly reduced the RNA-binding ability, whereas that at motif II did not cause a significant change.
Fig. 3.
The relative RNA-binding strength of variant BaMV helicase-like domains. (A) Northwestern assay. Proteins resolved on SDS-PAGE were transferred to nitrocellulose and allowed to refold in buffer as described in Materials and methods. After incubation with 32P-labeled RNA for 60 min, the membrane was visualized with a phosphorimager. The relative ability to bind the RNA probe in comparison to WT is indicated after normalization to the loaded protein. (B) Gel mobility shift assay. Different amounts (50, 130, or 260 ng) of the indicated protein were incubated with a 50-nt 32P-labeled RNA (50 ng) at 20 °C for 1 h prior to electrophoretic separation on 8% PAGE as described in Materials and methods. “−” indicates the migration of RNA probe in the absence of protein.
Mutational effects on ATPase and RTPase activities
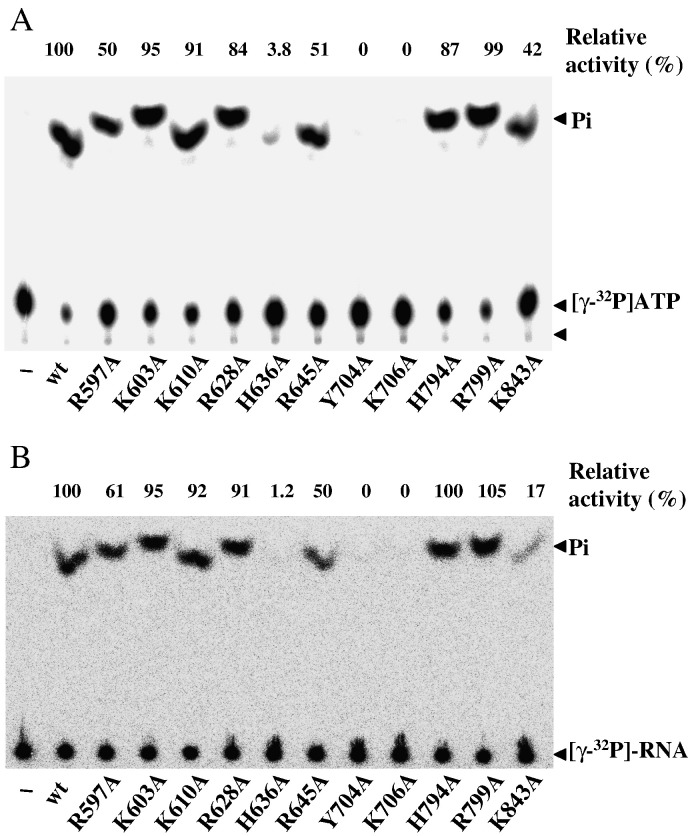
The importance of the selected residues in the release of 5′-γ-phosphate from ATP (Fig. 4A) and RNA (Fig. 4B) was assayed. Mutations of Y704 or K706, both located in motif II, abrogated both activities, suggesting that the critical region of motif II extends beyond the consensus DE/D residues. Mutation of H636, which is just in front of motif I, also severely inhibited activity. Substitution of alanine for R645, immediately downstream of the GKS signature of motif I, still retained approximately 50% activity. The mutational effects of R597A were around the same magnitude as R645A. K843A differentially affected ATPase and RTPase with the former decreased to 42% and the latter to 17% of WT. The rest of the mutant proteins were comparable to WT for both activities. The similar effects on ATPase and RTPase of mutations corroborate a previous hypothesis that the catalytic sites of the two activities overlap extensively.
Fig. 4.
Effects of mutations on ATPase and RTPase activities. (A) ATPase. Each reaction contained 30 ng indicated protein and 0.2 mM ATP (plus 1 μCi [γ-32p]ATP) in a final 3 μl buffer solution. The reaction was performed at 20 °C for 1 min. The reaction products were analyzed by TLC and visualized by autoradiography. (B) RTPase. The conditions were as in part A except that 0.4 μg [γ-32p]RNA was used as the substrate. The relative activity was estimated according to the pixels of released phosphates by considering WT as 100%.
Mutational effects on viral accumulation in vivo
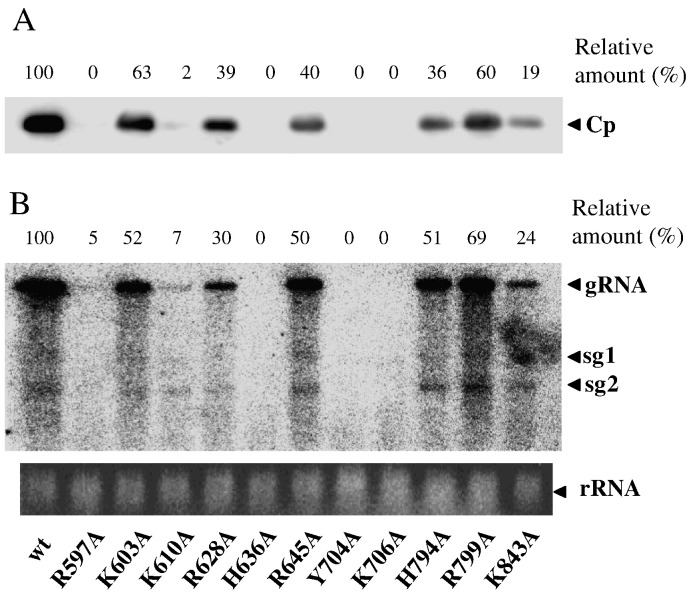
The involvement of the potential RNA-binding residues in viral replication was then examined by introducing pCBG, or its mutant derivatives, into protoplasts of Nicotiana benthamiana and measuring accumulation of the viral coat protein (Fig. 5A) and genomic RNA (Fig. 5B). In general, effects on the two measurements in response to a given mutation were similar, suggesting that the changes in coat protein accumulation mainly resulted from changes in the efficiency of viral replication. Virus carrying H636A, Y704A, or K706A failed to accumulate viral RNA and coat protein above detectable levels. The lack of the 5′ cap, due to the disabled RTPase, is enough to cause failure of the virus. By contrast, viruses with K603A, R628A, H794A, or R799A were able to reproduce, at an efficiency within ∼ 35–65% of WT, as their ATPase and RTPase activities were approximately equal to that of WT.
Fig. 5.
BaMV replication in protoplasts of N. benthamiana. (A) Accumulation of viral coat protein. Protoplasts (3 × 105) were inoculated with 2 μg pCBG or its derivative as indicated and 6 μg carrier DNA and incubated at room temperature for 48 h. The protein extracts (5 μg each) of the protoplasts were then analyzed by Western blot using anti-coat protein antibodies and chemiluminescence reagent. The relative amounts of the viral coat protein were estimated according to the luminescent signals by referring WT as 100%. (B) Accumulation of positive strand viral RNAs. Total RNA (0.5 μg) from protoplasts infected with pCBG or the indicated derivative was analyzed by Northern blot using a 0.6 kb RNA probe complementary to the 3′-terminal region of the genomic RNA. The viral genome (gRNA) and two subgenomic RNA (sg1 and sg2) are indicated. The relative amounts of the genomic RNA were estimated according to the respective pixels by considering WT as 100%.
Surprisingly, K610A could only replicate to a very limited extent, although it had > 90% enzymatic activities of WT. R597A and R645A both retained approximately half of the enzymatic activity of WT. R597A barely reproduced in protoplasts, however R645A could reproduce with efficiency consistent with its ATPase and RTPase activities. K843A mutation had similar effects on RTPase activity and replication efficiency, suggesting that RTPase activity as low as 20% of WT is enough to sustain viral replication. A previous study using the Q826A mutation suggested that greatly enhanced ATP-binding ability might deteriorate viral RTPase activity in vivo due to the mutual exclusion of the two substrates (Han et al., 2007). Could this also be an explanation for the unexpected results of the R597A and K610A mutations? Kinetic analysis of ATPase activity based on an enzyme-coupled assay was selectively carried out on WT, R597A, K603A, and K610A (Table 2 ). The magnitudes of K m and k cat were not changed significantly due to the mutations. Collectively, the results implied that in addition to its involvement in ATPase and RTPase activities, the BaMV helicase-like domain may have other functions required for replication.
Table 2.
Kinetic constants of various BaMV helicase-like domains toward ATP.
| Km (μM) | kcat (s− 1) | |
|---|---|---|
| WT | 148 ± 5 | 82 ± 1 |
| R597A | 137 ± 6 | 49 ± 1 |
| K603A | 161 ± 11 | 61 ± 2 |
| K610A | 126 ± 6 | 58 ± 2 |
The helicase-like domain was previously reported to have preferential binding to the rABC structural domain of the viral 3′UTR based on a UV-induced crosslinking (Chen et al., 2003). This specific protein–RNA interaction has been proposed to account for the structural requirement of the rABC domain for an efficient replication of BaMV. Thus, binding of the three mutant proteins to the 3′UTR was assayed with the same technique in this study (Fig. 6 ). These results showed that R597A, K603A, K610A, and WT exhibited similar binding strength to the 3′UTR, suggesting that this specific interaction is not the basis for the replication incompetence of R597A and K610A.
Fig. 6.
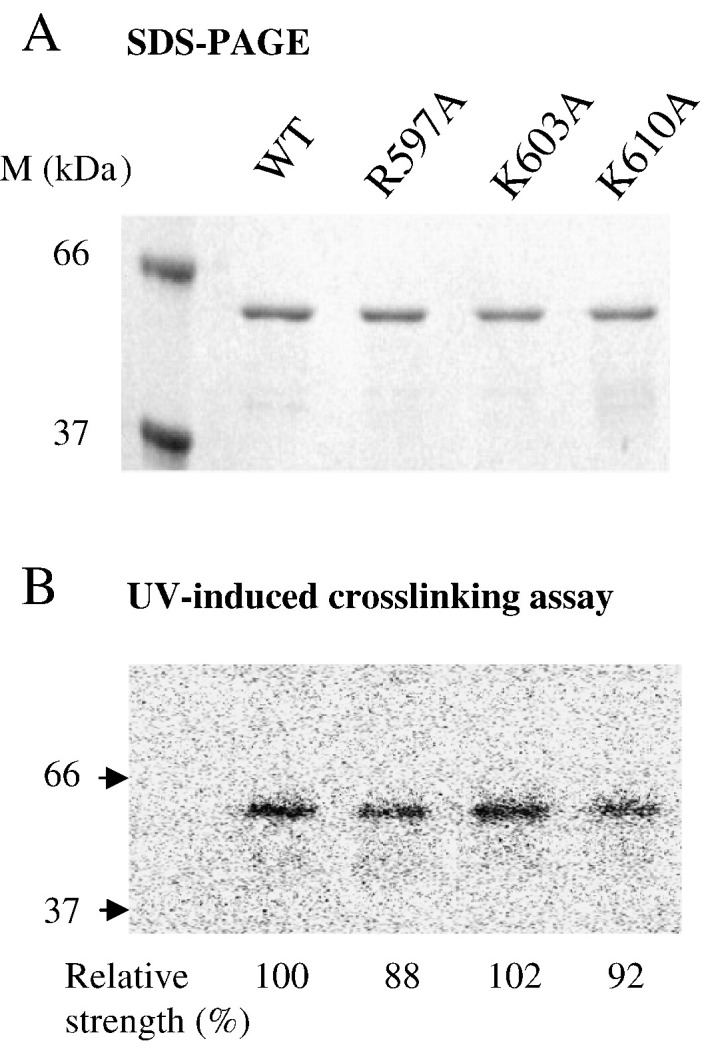
Affinity of variant BaMV helicase-like domains for 3′UTR. (A) Purity of the assayed proteins was examined by SDS-PAGE. (B) UV-induced crosslinking assay. The indicated protein was incubated with a 32P-labeled probe, which contains the rABC domains of the 3′UTR, for 10 min and irradiated by UV under conditions as described in Materials and methods. After treatment with RNase, the crosslinked proteins were resolved on SDS-PAGE and visualized with a phosphorimager. The relative ability to bind the RNA probe in comparison to WT is indicated after normalization to the loaded protein.
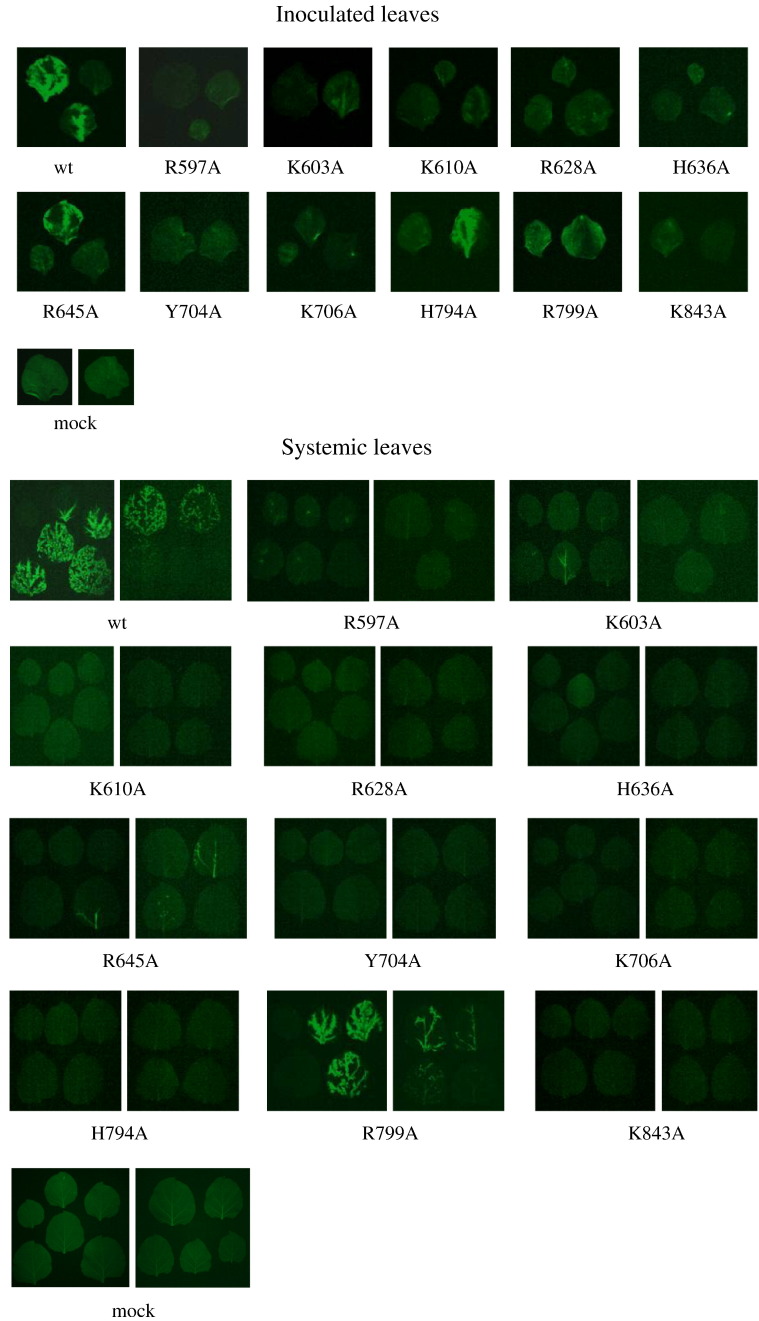
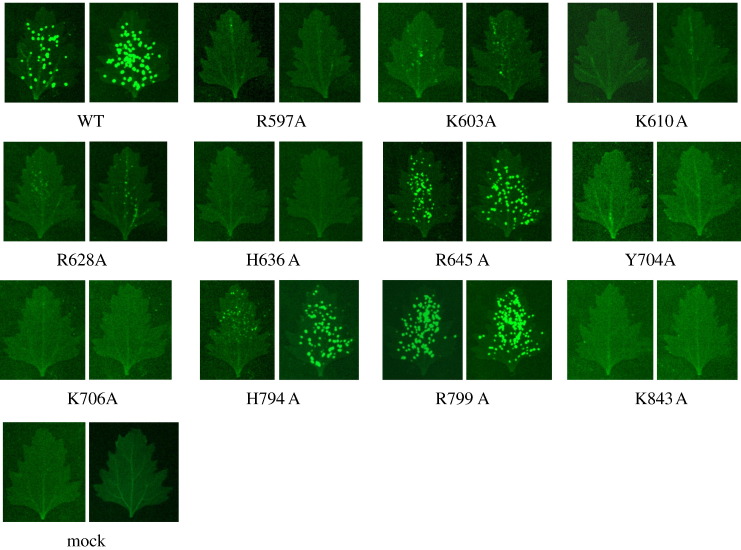
The influence of the mutated residues in viral infectivity was examined by inoculating the corresponding pCBG derivatives into leaves of N. benthamiana. The expression of GFP in inoculated and systemic leaves was used as an index for viral replication in plant (Fig. 7 ). With respect to inoculated leaves, inoculation with WT gave high GFP expression after 20 days post-inoculation (dpi). Sufficient GFP expression was also observed with R645A, H794A, and R799A. By contrast, only limited fluorescence was observed with K603A, R628A, and K843A. Furthermore, GFP expression was very restricted in response to R597A, K610A, H636A, Y704A, and K706A mutations. GFP accumulation in systemic leaves at 20 dpi was also recorded, which showed that only WT and R799A spread significantly. Infectivity of the BaMV variants in Chenopodium quinoa was also examined (Fig. 8 ). Many fluorescent spots appeared on the leaves infected by WT, R645A, H794A, and R799A at 7 dpi. By contrast, the fluorescent spots on leaves inoculated with K603A, and R628A were few and dim. The rest of the mutants did not give rise to notable GFP expression.
Fig. 7.
Fluorescent images of the leaves of N. benthamiana inoculated with various BaMV cDNA clones. The inoculation conditions are described in Materials and methods. The images were taken at 20 dpi. Systemic leaves for any given viral inoculation were from the same plant and placed (from old to young) in order from left to right, top to bottom.
Fig. 8.
Fluorescent images of the leaves of C. quinoa inoculated with various BaMV cDNA clones. The images were taken at 7 dpi.
Discussion
The majority of helicases can be categorized into three superfamilies based on the presence of defined helicase motifs. The GKS/T signature of motif I is highly conserved across all the three superfamilies, whereas the consensus sequences of motif II are slightly different, with DE/D in SF1 and SF3 and DEAD, DEAH, or DExH in different subfamilies of SF2. Other motifs including III, V, and VI are specific to each superfamily or subfamily. Comparison between the structures of SF1 helicases, e.g. Rep bound to ss-DNA (Korolev et al., 1997) and PcrA (Subramanya et al., 1996), and SF2 helicase, e.g. HCV NS3 (Yao et al., 1997), revealed that the core domains of the helicases have similar folds, characteristic of a large central parallel β-sheet flanked by α-helices, although there is little sequence homology between the SF1 and SF2 helicases (Korolev et al., 1998). Together with structures of other RNA-bound helicases, including eIF4AIII (Andersen et al., 2006) and DEAD-box protein Vasa (Sengoku et al., 2006), the functions of helicase motifs have been concluded as follows: motifs I, II, and VI for ATP binding and hydrolysis, motifs III and V for coordinating nucleic acid binding and ATPase activity, and the less conserved motifs including motif IV and motifs Ia, Ib, Ic, Id, IVa, IVb, and Va, when present, for nucleic acid binding (Lee and Yang, 2006, Jankowsky and Fairman, 2007).
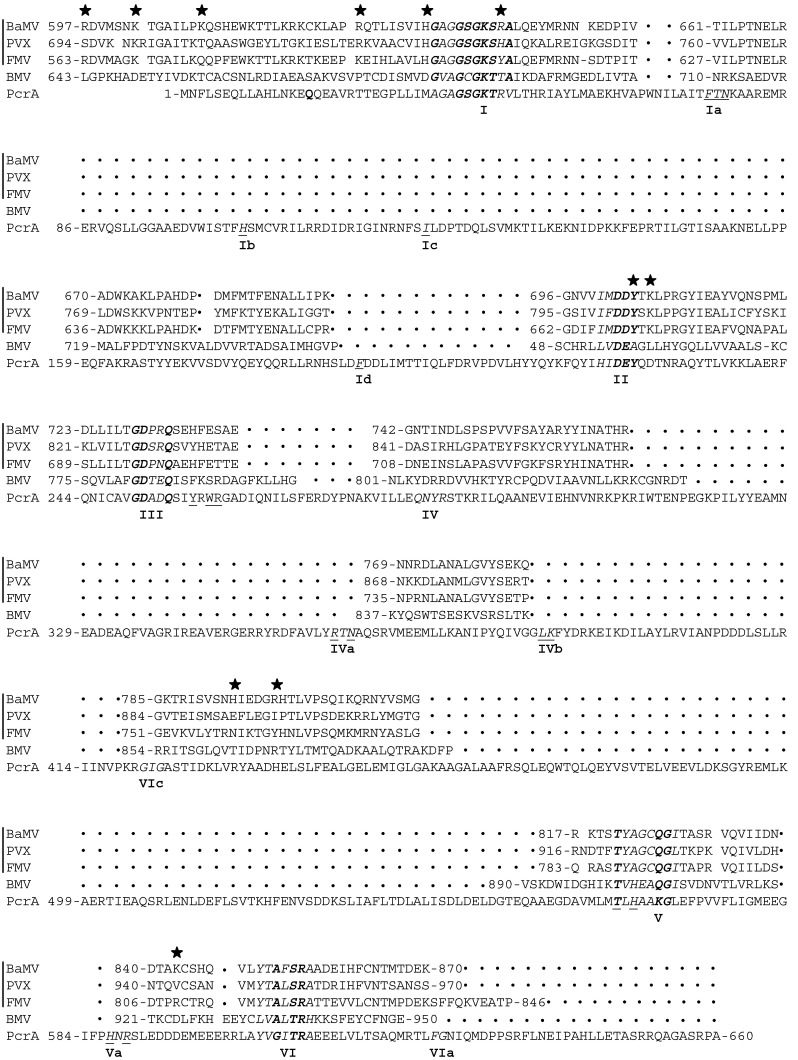
The helicase-like domain of BaMV replicase possesses NTPase and RTPase activities (Li et al., 2001). It is known that motifs I, II, and VI are essential for both of the activities (Han et al., 2007); however, the regions responsible for RNA binding remained largely unclear before this study. In the present study, formaldehyde crosslinking between the viral helicase-like domain and RNA identified several peptidyl regions that could potentially contact RNA. The finding of motif I- and II-containing regions in association with RNA is understandable, as the two motifs play essential roles in cleaving the 5′ γ-phosphate off the RNA substrate. Three other non-conserved peptidyl regions were also found to bind RNA. Compared with the bona fide helicases described above, the BaMV helicase seems to bind RNA by using different peptidyl regions. Specifically, the sequence of the BaMV helicase-like domain, along with several homologues from other plant viruses, aligns with the well-characterized DNA helicase PcrA (Fig. 9 ). As members of SF1, all the proteins have similar consensus sequences within motifs I, II, III, V, and VI. However, the helicase-like domains of plant viruses are shorter than PcrA and lack motifs Ia, Ib, Ic, Id, IVa, IVb, and Va, which are responsible for DNA binding in PcrA (Lee and Yang, 2006).
Fig. 9.
Alignment of some proteins belonging to helicase superfamily 1. The helicase motifs are shown in italic with consensus residues marked in bold. BaMV, PVX, FMV, and BMV represent the helicase-like domains from Bamboo mosaic virus, Potato virus X, Foxtail mosaic virus, and Brome mosaic virus, respectively. PcrA is a DNA helicase from Bacillus stearothermophilus. The residues selected for mutagenesis in this study are marked with asterisks. The residues involved in the binding of PcrA to nucleic acids are underlined. BaMV, PVX, and FMV are members of the potexvirus genus, while BMV belongs to bromovirus.
The importance of the mapped peptidyl regions in the enzymatic activities and viral replication was assayed by substituting alanine for the arginine, lysine, histidine, and tyrosine residues within the regions based on the preferential reactivity of these amino acids to formaldehyde and ability to link RNA. Eleven mutant proteins were thus created and purified. The capability of the BaMV helicase-like domain to bind RNA was not significantly affected by any single mutation. Similar results had been observed in a previous HCV NS5B-RNA binding study, in which a less than two-fold decrease in the affinity for RNA was seen in response to single point mutation (Kim et al., 2005). In addition, the magnitudes of K m of mutant proteins, R597A, K603A, and K610A, toward ATP were not altered significantly. Collectively, these results suggest that the mutations should not cause significant changes in the structure of BaMV helicase-like domain. Therefore, the effects on viral replication in vivo should be due to the inherent functions of the mutated residues, rather than the result of drastic changes of the protein structure.
The mutational effects can be categorized into several classes (Table 3 ). First, H636A, Y704A, and K706A caused severe or even deadly effects on not only in vitro ATPase and RTPase activities but also viral replication in protoplasts. Disabled enzymatic activities should be the cause for infertility of the mutant viruses in plant cells. In theory, RNA helicase activity is important for viral survival by unwinding duplex RNA, e.g. stem-loop and hairpin structures, to facilitate the replication process or avoid RNA interference. Although the possession of helicase activity remains uncertain, ATPase activity is a prerequisite for such a presumable activity. Without the fuel provided by ATPase, the presumable helicase activity could not function normally. RTPase is another essential function for viral replication. If unable to form the 5′-cap at plus sense viral RNAs, viral protein translation would be shut down. Conservation of these three residues in most potexviruses may provide a clue to the importance of the residues (Fig. 9).
Table 3.
Mutational effects of the BaMV helicase on various biological activities.
| Mutation | ATPase (%) | RTPase (%) | RNA binding (%) | Replication in protoplast (%) |
Infectivity in plants |
||
|---|---|---|---|---|---|---|---|
| gRNA | CP | Cell-to-cella | Systemicb | ||||
| WT | 100 | 100 | 100 | 100 | 100 | +++++ | +++++ |
| H636A | 3.8 | 1.2 | 81 | 0 | 0 | − | − |
| Y704A | 0 | 0 | 86 | 0 | 0 | − | − |
| K706A | 0 | 0 | 89 | 0 | 0 | − | − |
| R597A | 50 | 61 | 88 | 5 | 0 | − | − |
| K610A | 91 | 92 | 94 | 7 | 2 | − | − |
| K603A | 95 | 95 | 102 | 52 | 63 | + | + |
| R628A | 84 | 91 | 80 | 30 | 39 | + | − |
| K843A | 42 | 17 | 99 | 24 | 19 | − | − |
| R645A | 51 | 50 | 87 | 54 | 40 | ++++ | ++ |
| H794A | 87 | 100 | 83 | 51 | 36 | +++++ | + |
| R799A | 99 | 105 | 80 | 69 | 60 | +++++ | ++++ |
+++++: > 5 leaves contain GFP signal and < 3 leaves filled with GFP signal.
++++: > 5 leaves contain GFP signal.
+++: > 3 leaves contain GFP signal.
++: > 2 leaves contain GFP signal.
+: < 1 leaves contain GFP signal.
−: no leaves contain GFP signal.
The viral cell-to-cell movement in plants was determined by the number and size of GFP signal spots on C. quinoa leaves.
The viral systemic movement was estimated by the number of upper leaves showing GFP signal and the extent of GFP expression in N. benthamiana.
The second class includes R597A and K610A. Compared with the results of R645A and K843A, the remaining ATPase and RTPase activities of R597A and K610A should be able to support viral replication in protoplasts proportionally. However, replication of the two mutant viruses in protoplasts was very limited. This discrepancy suggests that the peptidyl region encompassing residues 597–610 may be involved in the unproven helicase activity or some other unidentified functions required for replication. BMV 1a protein represents an example that demonstrates the involvement of a helicase-like domain in viral replication by additional mechanisms such as assembling the replication complex and regulating viral protein translation (Chen and Ahlquist, 2000, Chen et al., 2001, Yi et al., 2007).
The third class includes K603A and R628A, which limited viral infectivity in plants despite some amount of viral replication in protoplasts. Accumulations of the coat protein and genomic RNA of the two mutant viruses in protoplasts were approximately 30–63% of WT. Similar levels of accumulation also resulted from R645A, H794A, and R799A. However, there is an apparent disparity in viral infectivity between the two groups. The former accumulated much lower amounts of GFP than the latter in the inoculated leaves of C. quinoa and N. benthamiana, suggesting that viral cell-to-cell movement was somewhat restricted by K603A and R628A mutations. The involvement of the helicase-like domain in viral cell-to-cell movement was also demonstrated previously in Tobacco mosaic virus (Hirashima and Watanabe 2003). Mutations of R645A and H794A constitute the fourth class. Although virus carrying either mutation could reproduce properly in inoculated leaves of N. benthamiana, it could not move into systemic leaves as efficiently as WT and R799A. R799A belongs to another class. No adverse effect regarding replication and movement was noted due to the R799A mutation.
With RNA-binding ability, helicases have evolved diverse functions to cope with the demands for the organism survival. For example, the RNA helicases Ded1 and NPH-II can displace protein from RNA in an ATP-dependent mode without duplex unwinding (Fairman et al., 2004, Jankowsky and Bowers, 2006). It is tempting to assume that such a protein-displaced activity would be useful for BaMV to control the opposite movements between ribosome and replicase complex along the viral RNA. Cellular DExH proteins RIG-I and MDA5 can even sense the presence of viral RNA in cytoplasm and initiate antiviral cellular responses (Pichlmair et al., 2006, Yoneyama et al., 2004 , 2005). With the potential versatility of helicases, it is not surprising to discover that the BaMV helicase-like domain may be involved in various biological functions, including 5′-cap formation, replication, and viral movement. This study thus provides a base for future investigation on each of the mechanisms.
Materials and methods
Plasmids and site-directed mutagenesis
The plasmid pHWT was used as an expression vector for the BaMV helicase-like domain in Escherichia coli Novablue (DE3) cells as described previously (Li et al., 2001). Plasmid pCBG is an infectious clone of recombinant BaMV in which the green fluorescent protein (GFP) gene preceded by a duplicated coat protein promoter is inserted between triple gene block and coat protein-coding regions as described previously (Han et al., 2007). pUHel is a pUC18-based vector containing a SphI-digested fragment (4168 nts) isolated from pCBG. Point mutations in pHWT and pUHel were created according to the protocol of the QuikChange site-directed mutagenesis kit (Stratagene). All mutations were confirmed with an ABI Prism 3100 auto sequencer (PerkinElmer). After obtaining the desired mutations, the mutated SphI-digested fragment in pUHel was cloned back into pCBG.
Protein expression and purification
The culture for inducing protein expression in E. coli and the initial purification, including steps for dissolution of inclusion bodies, protein refolding, and immobilized metal affinity chromatography, were performed as described previously (Li et al., 2001, Han et al., 2007). Gel filtration chromatography using Hiprep 16/60 Sephacryl S-300 (Pharmacia) was included to further separate the refolded proteins based on their oligomerization states. The purified protein was finally stocked in 50 mM Tris [pH 7.5], 150 mM KCl, 0.1% Brij-35, 10% glycerol, and 5 mM β-mercaptoethanol. Thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa) were used in gel filtration chromatography as markers for the molecular weight calibration of the eluted proteins.
RNA preparation
A double-stranded DNA template containing a T7 promoter followed by 15 nt similar to the 5′ sequence of BaMV genomic RNA was prepared by annealing 5′-TAATACGACTCACTATAGGGAAACCACGCCAT and 5′-ATGGCGTGGTTTCCCTATAGTGAGTCGTATTA oligonucleotides. One μg template was then used to produce a 15-nt RNA with a biotin molecule attached at the 3′ end in a 20-μl in vitro transcription reaction that also contained 3.75 mM ATP, 3.75 mM GTP, 3.75 mM CTP, 2 mM Bio-16-UTP (Ambion), 2 μl T7-MEGAshortscript enzyme mix (Ambion), and 1 × T7 transcription buffer. A control reaction with the same buffer except for the omission of Bio-16-UTP was performed in parallel. The biotin-labeled RNA was identified and recovered from a 15% acrylamide–7 M urea gel using RNA products produced in the control reaction as references.
A 5′-[γ-32P]-labeled transcript corresponding to the first 50 nt of genomic RNA was used in the RTPase assay as described previously (Han et al., 2007). A 5′-terminally 178 nt genomic transcript, body-labeled with 32P, was prepared for Northwestern assay. A 32P-labeled 600 nt transcript complementary to the 3′ end of the genomic RNA was prepared for the detection of genomic RNAs in Northern blot assay as described previously (Huang and Tsai, 1998). Another 32P-labeled 100 nt RNA transcript, containing the rABC domains of the 3′UTR, was synthesized for use in the UV-induced crosslinking assay as previously described (Chen et al., 2003).
Enzymatic activity assay
ATPase and RTPase activities were determined by TLC based on the release of 32P from [γ-32P]ATP and 5′-[γ-32P]RNA, respectively (Li et al., 2001). ATPase activity was also determined by an enzyme-coupled assay in which ATP hydrolysis was linked to NADH oxidation through the activities of pyruvate kinase and lactate dehydrogenase (Han et al., 2007). Kinetic constants of ATP hydrolysis were calculated by Grafit software using data generated from the enzyme-coupled assays.
Reversible formaldehyde crosslinking of protein and biotinylated RNA
The procedure for crosslinking the helicase-like domain to biotinylated RNA by formaldehyde was based on a previously-published method (Kim et al., 2005). Briefly, 2 μM biotinylated RNA and 1 μM protein were incubated together in 10 mM Hepes [pH 7.5], 75 mM KCl, and 0.2% (v/v) formaldehyde at room temperature for 10 min. The reaction was quenched by adding glycine to a final concentration of 0.2 M. The crosslinked complex was precipitated by mixing with an equal volume of acetone at − 20 °C for 60 min and subjected to centrifugation at 15,000 rpm for 60 min at 4 °C. The dried pellet was then dissolved in 50 μl 100 mM ammonium bicarbonate [pH 8.0]. Sequencing grade trypsin (Trypsin Gold, Promega) was added in a ratio of protease:substrate of 1:10 (w/w), and the digestion was performed at 37 °C overnight. TetraLink tetrameric avidin resin (Promega), equilibrated with 25 mM ammonium bicarbonate [pH 7.8], was used to capture the biotinylated RNA–peptides complex. After extensive washing, at least six times, with the same buffer, 50 μl of 0.5% (v/v) trifluoroacetic acid was added, and the mixture was incubated at 70 °C for 60 min to reverse the crosslink between the RNA and peptides. The released peptide in the supernatant was recovered after a 5 min centrifugation at 10,000 rpm and dried in a speed vacuum. After being dissolved in 0.1% formic acid, the sample was purified with Ziptip (Millipore). A parallel treatment without the addition of formaldehyde was carried out as a background control. The peptide identification was performed using an Applied Biosystems QStar LC-MS/MS spectrometer equipped with LC system. The obtained mass spectrometry information was analyzed with Mascot software (Matrix Science).
RNA-binding assay
The Northwestern experiment was modified from Robbins et al. (1986). Briefly, proteins resolved on a polyacrylamide gel were first transferred to a nitrocellulose paper (0.45 μm, MILLIPORE), and the membrane was then incubated in 10 mM Tris [pH 7.0], 50 mM NaCl, 1 mM EDTA, 0.02% bovine serum albumin, 0.02% Ficoll, and 0.02% polyvinyl pyrrolidone for three times of 90 min each, and probed with approximately 1 × 106 cpm of 32P-labeled RNA for 60 min. Finally, the membrane was washed and dried. The bound RNA on the membrane was visualized with a phosphorimager (Fujifilm BAS 2500).
The gel mobility shift assay was carried out by incubating different amounts of the helicase-like domain (50, 130, or 260 ng) with 50 ng of 32P-labeled RNA probe, the first 50 nt of genomic RNA, in buffer that contained 50 mM Tris [pH 7.5], 100 mM KCl, 10% glycerol, 0.1% Brij-35, and 1 U RNase inhibitor at 20 °C for 1 h. The mixture was then separated on 8% PAGE and visualized with a phosphorimager.
The UV-induced crosslinking assay was performed by incubating indicated protein (140 ng each) with 5 pmol 32P-labeled RNA probe in a 10-μl buffer that also contained 10 mM Hepes [pH 7.4] and 10% (v/v) glycerol for 10 min at room temperature. The mixture was then irradiated on ice under 302 nm with energy of 120 mJ/sec/cm2 (Ultraviolet crosslinker, UVP) at a distance of 10 cm for 1 min. The irradiated products, after being digested with 1 μg RNase A at 37 °C for 2 h, were resolved on a 12% SDS-polyacrylamide gel and visualized with a phosphorimager.
Viral replication assay in protoplasts
Protoplast preparation from young leaves of N. benthamiana and the subsequent transfection were performed as the previous description (Tsai et al., 1999). In brief, each 3 × 105 protoplast was introduced with 2 μg pCBG (or its derivatives) and 6 μg carrier DNA, and the cells were then incubated at 25 °C for 48 h under constant illumination. The accumulation of coat protein in protoplasts was assayed by Western blotting analysis using the coat protein specific antibodies and chemiluminescence reagent (PerkinElmer). The luminescent signals were processed with a fluorimager (Kodak Image Station 2000MM). For viral RNA analysis, total RNA from protoplasts was extracted, glyoxalated, electrophoresed through 1% agarose gel, transferred to nylon membranes, and hybridized with a 32P-labeled RNA probe complementary to 3′UTR of BaMV as described earlier.
Viral infectivity assay in plants
Each of the selected leaves of 3-week-old N. benthamiana or C. quinoa (3–4 per plant) was inoculated with 2 μg pCBG (or its derivatives) in 10 μl aqueous solution by gently rubbing over the surface under the aid of sterile carborundum according to the method described previously (Lin and Hsu, 1994). Each plasmid DNA was applied to total six plantlets. Fluorescent images of the inoculated leaves and upper leaves were obtained at 20 dpi with a fluorimager using an excitation filter of 465 nm and an emission filter of 535 nm.
Acknowledgments
This work was supported by grants, NSC 95-2752-B-005-011-PAE and NSC 96-2752-B-005-011-PAE, from the National Science Council, Taiwan, Republic of China.
References
- Andersen C.B., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C.L., Pedersen J.S., Seraphin B., Le Hir H., Andersen G.R. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Bartelma G., Padmanabhan R. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002;299:122–132. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- Bernard M., Kersten G.F.A., Hoogerhout P., Brugghe H.F., Timmermans H.A.M., Jong A.D., Meiring H., Hove J.T., Hennink W.E., Crommelin D.J.A., Jiskoot W. Identification of formaldehyde-induced modifications in proteins. J. Biol. Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Bisaillon M., Lemay G. Characterization of the reovirus λ1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 1997;272:29954–29957. doi: 10.1074/jbc.272.47.29954. [DOI] [PubMed] [Google Scholar]
- Chen J., Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Noueiry A., Alquist P. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 2001;75:3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-H., Meng M., Hsu Y.-H., Tsai C.-H. Functional analysis of the cloverleaf-like structure in the 3′ untranslated region of Bamboo mosaic potexvirus RNA revealed dual roles in viral RNA replication and long distance movement. Virology. 2003;315:415–424. doi: 10.1016/s0042-6822(03)00560-9. [DOI] [PubMed] [Google Scholar]
- Fairman M.E., Maroney P.A., Wang W., Bowers H., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D RNA helicases without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Helicases: amino acid comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- Goregaoker S.P., Culver J.N. Oligomerization and activity of the helicase domain of the Tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 2003;77:3549–3556. doi: 10.1128/JVI.77.6.3549-3556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-T., Tsai C.-S., Chen Y.-C., Lin M.-K., Hsu Y.-H., Meng M. Mutational analysis of a helicase motif-based RNA5′-triphosphatase/NTPase from Bamboo mosaic virus. Virology. 2007;367:41–50. doi: 10.1016/j.virol.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-Y., Tsai C.-H. Evolution of Bamboo mosaic virus in a nonsystemic host results in mutations in the helicase-like domain that cause reduced RNA accumulation. Virus Res. 1998;58:127–136. doi: 10.1016/s0168-1702(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Hirashima K., Watanabe Y. RNA helicase domain of tobamovirus replicase executes cell-to-cell movement possibly through collaboration with its nonconserved region J. Virol. 2003;77:12357–12362. doi: 10.1128/JVI.77.22.12357-12362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., Meer Y.V.D., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Fairman M.E. RNA helicases — one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Kadaré G., Haenni A.L. Virus-encoded RNA helicases. J. Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-.C., Russell W.K., Ranjith-Kumar C.T., Thomson M., Russell D.H., Kao C.C. Functional analysis of RNA binding by the Hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:38011–38019. doi: 10.1074/jbc.M508145200. [DOI] [PubMed] [Google Scholar]
- Korolev S., Hsieh J., Gauss G.H., Lohman T.M., Waksman G. Major domain swivelling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Korolev S., Yao N., Lohman T.M., Weber P.C., Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases protein. Science. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M.K., Patel S.S. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- Li Y.-I., Shih T.-W., Hsu Y.-H., Han Y.-T., Huang Y.-L., Meng M. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′-triphosphatase activity. J. Virol. 2001;75:12114–12120. doi: 10.1128/JVI.75.24.12114-12120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N.-S., Hsu Y.-H. A satellite RNA associated with Bamboo mosaic potexvirus. Virology. 1994;202:707–714. doi: 10.1006/viro.1994.1392. [DOI] [PubMed] [Google Scholar]
- Lin N.-S., Lin B.-Y., Lo N.-W., Hu C.-C., Chow T.-Y., Hsu Y.-H. Nucleotide sequence of the genomic RNA of Bamboo mosaic potexvirus. J. Gen. Virol. 1994;75:2513–2518. doi: 10.1099/0022-1317-75-9-2513. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Perez-Romero P., Imperiale M.J. Assaying protein–DNA interactions in vivo and in vitro using chromatin immunoprecipitation and electrophoretic mobility shift assays. Methods Mol. Med. 2007;131:123–139. doi: 10.1007/978-1-59745-277-9_10. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis E., Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 50 phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Robbins S.G., Frana M.F., McGowan J.J., Boyle J.F., Holmes K.V. RNA-binding proteins of MHV: detection of monomeric and multimeric N protein with an RNA overlay-protein blot assay. Virology. 1986;150:402–410. doi: 10.1016/0042-6822(86)90305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- Tsai C.-H., Cheng C.-P., Peng C.-W., Lin B.-Y., Lin N.-S., Hsu Y.-H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for an efficient replication of Bamboo mosaic potexvirus RNA. J. Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann-Schiffler H., Seinsoth S., Stahl H. Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res. 2002;30:3192–3201. doi: 10.1093/nar/gkf416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Merits A., Auvinen P., Kääriäinen L. Identification of a novel function of the alphavirus capping apparatus: RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 2000;275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- Wang X., Lee W.-M., Watanabe T., Schwartz M., Janda M., Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 2005;79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N., Hesson T., Cable M., Hong Z., Kwong A.D., Le H.V., Weber P.C. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- Yi G., Gopinath K., Kao C.C. Selective repression of translation by the Brome mosaic virus 1a RNA replication protein. J. Virol. 2007;81:1601–1609. doi: 10.1128/JVI.01991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.-M., Gale M.J., Akira S., Yonebara S., Kato A., Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]