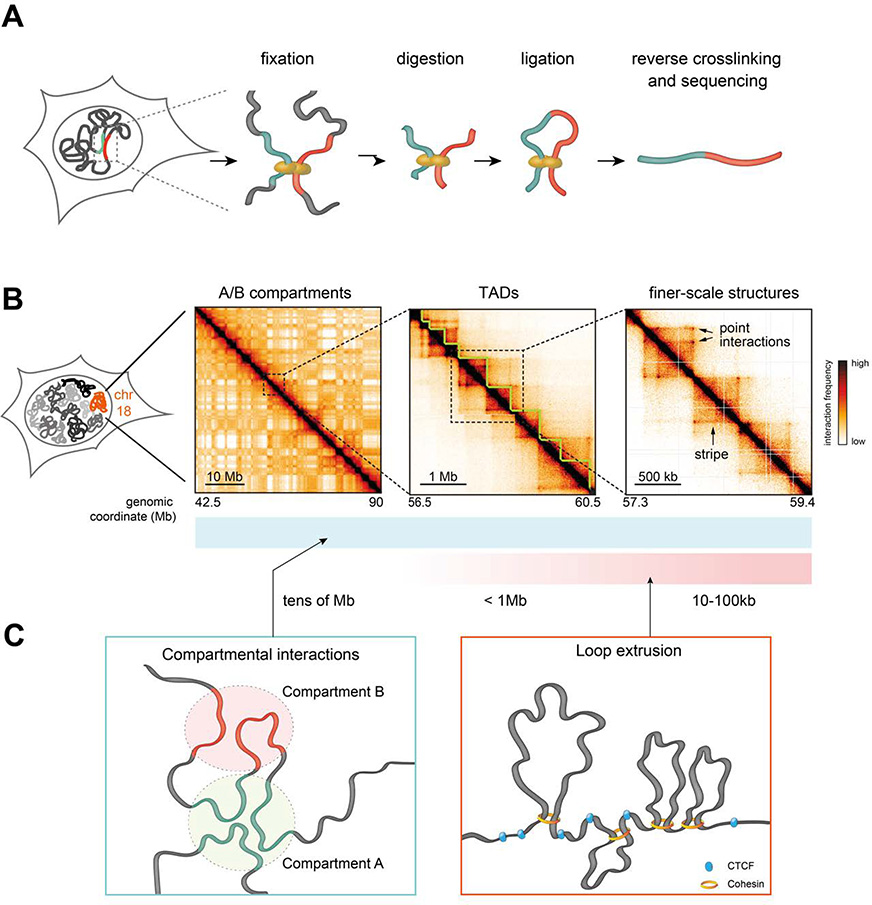
Figure 1.
A. Scheme of the core steps in chromosome conformation capture (3C) methods. Chromatin is crosslinked in cell nuclei and digested with a restriction enzyme (or endonuclease in the case of Micro-C), followed by ligation and decrosslinking. This results in the formation of hybrid DNA molecules that can be identified by high-throughput sequencing.
B. Hi-C contact maps illustrating that mammalian chromosomes are folded into checkerboard-like A/B compartments (left panel), TADs (middle panel), and shorter-scale structures. Sub-TAD structures include CTCF-related point interactions and stripes, as well as other interactions e.g. compartmental and polycomb-associated interactions (not shown in this example). Hi-C data were obtained mouse embryonic stem cells, from Ref. (Redolfi et al. 2019). Colormap saturation and scaling were modified across the three examples to emphasize structural features.
C. Hierarchies observed in chromosome folding are mainly driven by compartmental interactions involving attractions between active and inactive chromatin regions, manifesting at all genomic length scales; and CTCF/cohesin-mediated interactions likely originating from the loop extrusion activity of cohesin that is arrested by CTCF bound to DNA in defined orientations.