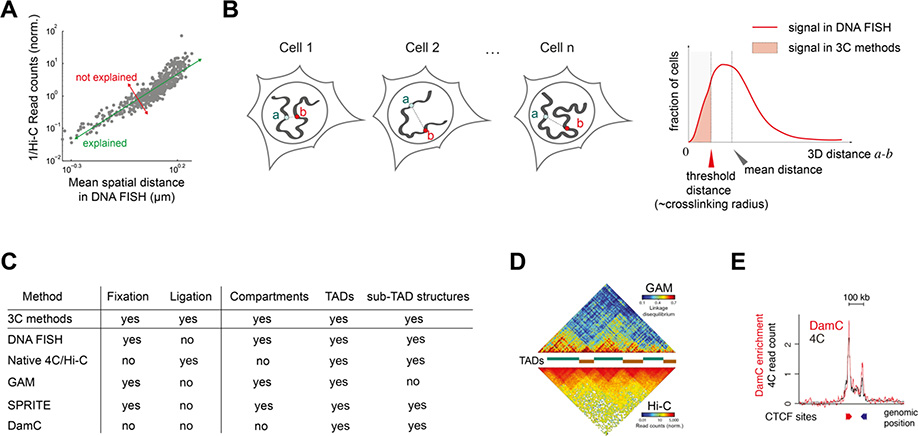
Figure 2.
A. 3C-based counts (Hi-C in this case) and spatial distances between loci measured in DNA FISH are generally well correlated, with a fraction of the variability that cannot be explained in terms of each other technique. Adapted from Ref. (Wang et al. 2016).
B. DNA FISH measures 3D distances between genomic loci (a and b here) and their distribution across the cell population. Signals in 3C methods arise from the fraction of cells where a and b can be crosslinked, which is usually (but not always) correlated with their average distance.
C. A summary of recently developed methods that are orthogonal to 3C and FISH and the structures they detected.
D. Ligation-free Genome Architecture Mapping (GAM) detects TADs and compartments in agreement with Hi-C. Adapted from Ref. (Beagrie et al. 2017)
E. Crosslinking and ligation-free DamC detects TAD boundaries and loops between convergent CTCF sites, in agreement with 4C-seq. Adapted from Ref. (Redolfi et al. 2019).