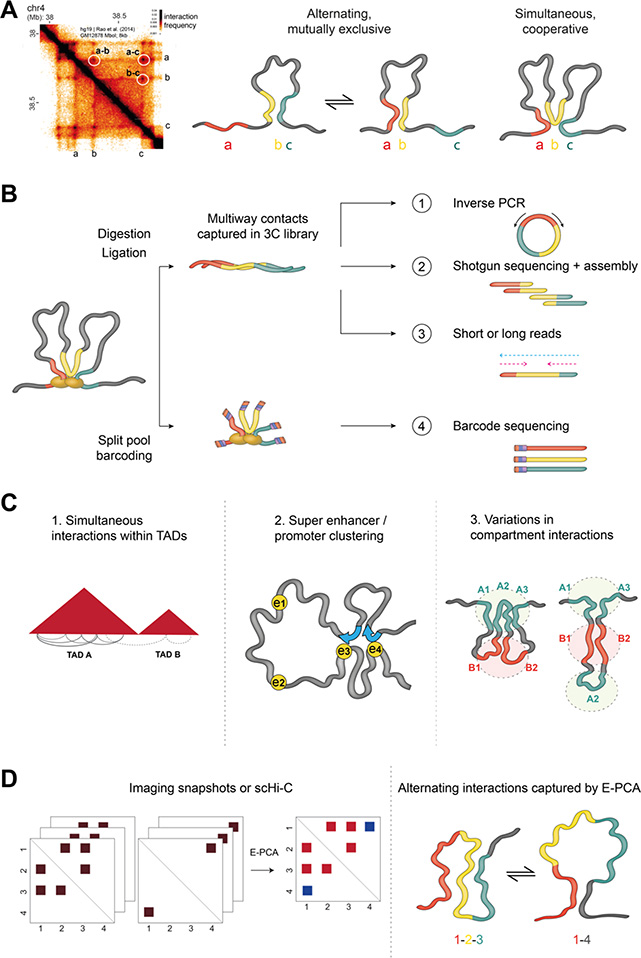
Figure 3.
A. Hi-C contacts between pairs of genomic loci (e.g. a, b and c here) can either correspond to mutually exclusive or to simultaneous (and possibly cooperative) interactions in single cells.
B. Simultaneous multi-way interactions can be identified using modified 3C methods that implement alternative strategies to sequence concatamers that exist in all 3C libraries, such as inverse PCR in MC-4C (scheme 1), shotgun sequencing and assembly in C-walks (2) or short-read sequencing as in Tri-C and similar methods (3). Multi-way contacts are also retrieved in SPRITE (4) using split-pool barcoding followed by sequencing of barcodes to identify genomic regions that were captured in the same cluster.
C. Multi-way chromosome conformation capture methods, as well as SPRITE and GAM, have shown overall that simultaneous contacts occur more frequently within TADs than across TAD boundaries (left), simultaneous interactions can occur between promoters and clusters of super-enhancers (middle), and that different subsets of A and B compartments cluster together in different subsets of cells (right).
D. E-PCA uses sets of contact matrices and reports patterns of correlated or anticorrelated contacts. Left: simplistic example where two possible contact patterns are found across single cells. Middle: The E-PCA result reports that the red set of interactions occur together and in opposition to the blue interactions, as in the schematic on Right.